Fig. 1.
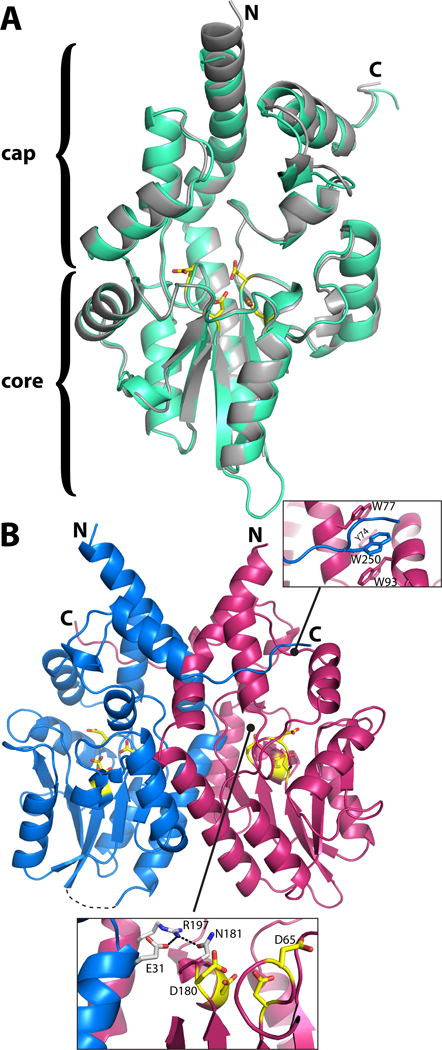
Structure of rPmCCAP. (A) Ribbon drawing of the protomer highlighting domain structure and similarity to rP4. rPmCCAP and rP4 (PDB code 3ET4) are shown in silver and green, respectively. Side chains of the four conserved Asp residues (DDDD motif) of rPmCCAP are shown in yellow. (B) Ribbon drawing of a dimer of rPmCCAP. The two chains are colored blue and pink. Asp residues of the DDDD motif are colored yellow. The dashed curve represents a disordered loop (residues 52–56) of chain A. The two insets show close-up views of intersubunit interactions.