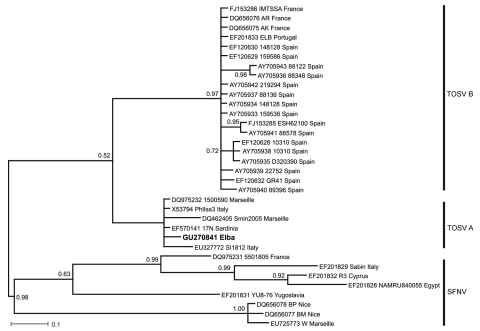
Figure.
Bayesian phylogenetic tree of Toscana virus (TOSV) and Sandfly fever Naples virus (SFNV) strains. For each sequence used, GenBank accession number, strain designation, and strain origin are shown. Phylognetic analysis was performed by using MrBayes 3.0 program (4) with a general time reversible substitution model. Substitution rates were assumed to follow a gamma plus invariants distribution. Three heated chains and a single cold chain were used in all Markov Chain Monte Carlo analyses, which were run for 1,000,000 generations, sampling 1 tree every 100 generations. Trees obtained before convergent and stable likelihood values were discarded (i.e., a 2,500 tree burn-in). Four independent runs, each started from different, randomly chosen trees, were performed to assess convergence. Posterior probabilities for nodes were assembled from all post burn-in trees (i.e., 30,004 trees per analysis). Posterior probabilities are shown on each node. Scale bar indicates nucleotide substitutions per site. The newly described TOSV sequence from Elba is shown in boldface.