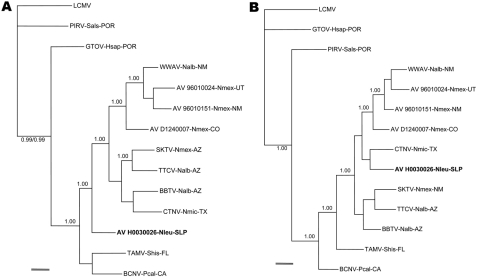
Figure 2.
Phylogenetic relationships among 11 North American arenaviruses based on Bayesian analyses of A) glycoprotein precursor gene sequences and B) nucleocapsid protein gene sequences. The number(s) at the nodes are clade probability values, a single 1.00 indicates that the clade probability values for both analyses were 1.00, and clade probability values <0.93 were not included in the phylograms. The branch labels include (in the following order) virus, host species, and state. BBTV, Big Brushy Tank virus strain AV D0390174 (GenBank accession No. EF619035); BCNV, Bear Canyon virus strain AV A0070039 (AY924391); CTNV, Catarina virus strain AV A0400135 (DQ865244); SKTV, Skinner Tank virus strain AV D1000090 (EU123328); TAMV, Tamiami virus strain W 10777 (AF512828); TTCV, Tonto Creek virus strain AV D0150144 (EF619033); WWAV, Whitewater Arroyo virus strain AV 9310135 (AF228063); arenaviruses AV 96010024 (EU123331), AV 96010151 (EU123330), and AV D1240007 (EU123329); GTOV, Guanarito virus strain INH-95551 (AY129247); PIRV, Pirital virus strain VAV-488 (AF485262); LCMV, lymphocytic choriomeningitis virus strain WE (M22138). AZ, Arizona; CA, California; CO, Colorado; FL, Florida; NM, New Mexico; SLP, San Luis Potosí; TX, Texas; UT, Utah; POR, Portuguesa (Venezuela). Nalb, Neotoma albigula (white-throated woodrat); Nleu, N. leucodon (white-toothed woodrat); Nmex, N. mexicana (Mexican woodrat); Nmic, N. micropus (southern plains woodrat); Pcal, Peromyscus californicus (California mouse); Sals, Sigmodon alstoni (Alston’s cotton rat); Shis, S. hispidus (hispid cotton rat). Pirital virus and GTOV are South American Tacaribe serocomplex viruses and were selected to represent South American lineages A and B, respectively. The LCMV strain WE is a member of the lymphocytic choriomeningitis-Lassa (Old World) serocomplex and was included in the analyses to enable inference of the ancestral node among the North American arenaviruses. Scale bars indicate 0.1 substitutions per site.