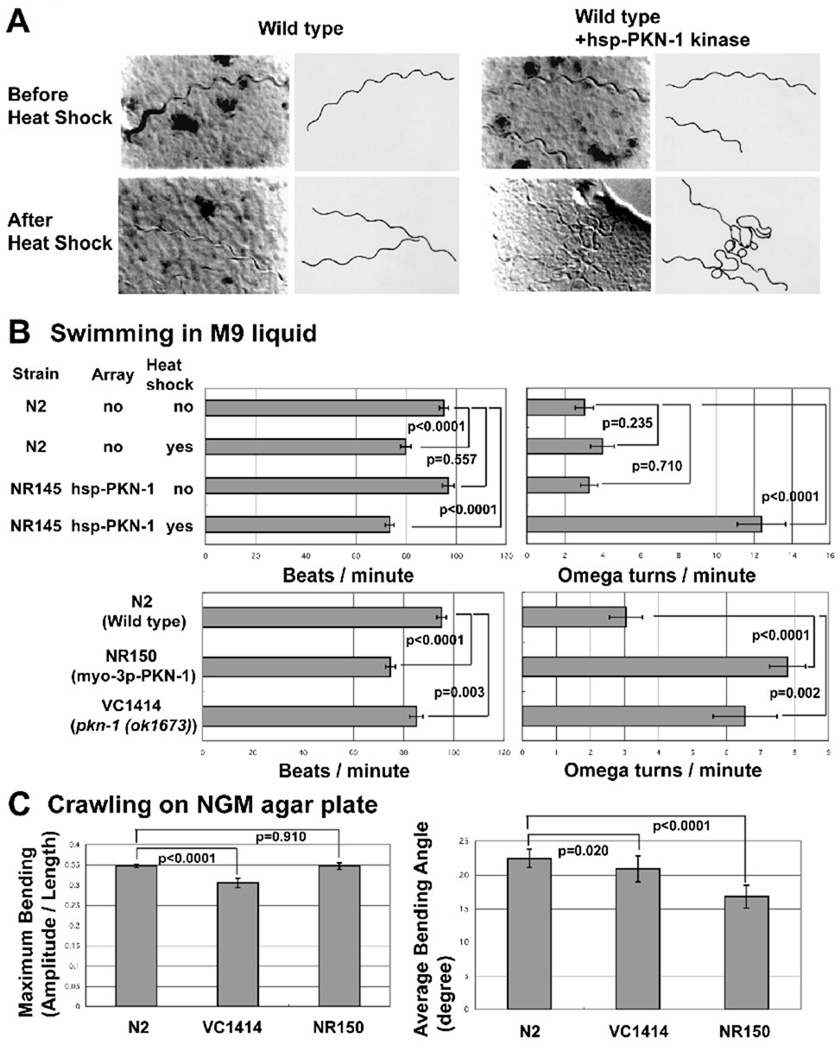
Figure 4. Either gain of function or loss of function of pkn-1 results in a “loopy Unc phenotype”.
A: Images (left) together with tracings (right) of worm tracks made along a bacterial lawn, from wild type, or wild type with hsp (heat shock promoter)-PKN-1 kinase before or after heat shock (30°C for 2 hrs). Without heat shock, wild type and wild type with hsp-PKN-1 kinase domain worms showed normal movement. After heat shock, only wild type with hsp-PKN-1 kinase domain worms showed more loopy tracks on the bacterial lawn surface. Transgenic worms harboring an extrachromosomal array consisting of hsp-PKN-1 kinase domain and sur-5::nls::gfp was constructed by injection34 of DNA mixture of pPD49.78-PKN-1 kinase, pPD49.83-PKN-1 kinase, and pTG96 (sur-5::nls::gfp marker plasmid) into N2 wild type, and selecting progeny showing GFP expression. The extrachromosomal array was integrated by UV irradiation35. To construct pPD49.78-PKN-1 kinase and pPD49.83-PKN-1 kinase, PCR amplified pkn-1 cDNA containing the kinase domain (see Figure 1A) was cloned into pPD49.78 or pPD49.83.
B: Bar graphs showing the results of two assays of adult worm swimming in M9 liquid: beats per minute (left) and omega turns per minute (right), each with means and standard errors (SEs) (n=20). Pair-wise comparisons for statistical significance are indicated with brackets and p values. These comparisons were made with one-way ANOVA using the post hoc Tukey HSD test, on StatPlus (AnalystSoft, Inc.). NR145 (harboring hsp-PKN-1 kinase) with heat shock, NR150 (harboring myo-3p-PKN-1 kinase), VC1414 (pkn-1 genomic deletion) show increased omega turn frequency. NR150 and VC1414 show reduced beats per minute. NR150 is an integrated line of the myo-3p-PKN-1 kinase array. The myo-3 promoter is reported to be expressed only in muscle36. To construct the plasmid expressing the PKN-1 kinase domain under the control of the myo-3 promoter (pPD95.86-PKN-1 kinase), a PCR amplified cDNA fragment for pkn-1 kinase domain was cloned into pPD95.86. Transgenic worms harboring extrachromosomal arrays were obtained by injection34 of myo-3p-PKN-1 kinase plasmid with pTG96 (sur-5::nls::gfp) into wild type worms. The extrachromosomal array was integrated by UV irradiation35. We obtained VC1414 from Caenorhabditis Genetics Center.
C: Bar graphs showing the maximum bending amplitude during an induced reversal (left) and the average bending angle during normal forward locomotion (right) of N2 (wild type), VC1414 (pkn-1 genomic deletion), and NR150 (integrated myo-3p-PKN-1 kinase), each with means and SEs. For maximum bending amplitude, n=20 except for N2 (n=65). For average bending angle, n=15 for each strain. Pair-wise comparisons for statistical significance are indicated with brackets and p values. These comparisons were made with one-way ANOVA using the post hoc Bonferroni test for the graph on the left (Maximum Bending), the post hoc Tukey HSD test for the graph on the right (Average Bending Angle), on StatPlus (AnalystSoft, Inc.). Each assay was performed on a blank NGM agar plate without bacteria. The average bending angle is the average of the absolute values of the angles of the worm when it is divided into 12 equal segments (13 points). The angle is between consecutive segments and relative to 180 degrees. The deeper the bend, the higher the angle. The numbers shown are averages over a 6–15 second interval when the animals were moving forward. VC1414 shows a reduction of maximum bending ability. NR150 shows a reduction of average bending angle. The methods for these measurements were described previously22, 37, 38.