Zds1/2 regulate mitotic progression by directing the nucleocytoplasmic distribution of Cdc55–PP2A.
Abstract
Budding yeast CDC55 encodes a regulatory B subunit of the PP2A (protein phosphatase 2A), which plays important roles in mitotic entry and mitotic exit. The spatial and temporal regulation of PP2A is poorly understood, although recent studies demonstrated that the conserved proteins Zds1 and Zds2 stoichiometrically bind to Cdc55–PP2A and regulate it in a complex manner. Zds1/Zds2 promote Cdc55–PP2A function for mitotic entry, whereas Zds1/Zds2 inhibit Cdc55–PP2A function during mitotic exit. In this paper, we propose that Zds1/Zds2 primarily control Cdc55 localization. Cortical and cytoplasmic localization of Cdc55 requires Zds1/Zds2, and Cdc55 accumulates in the nucleus in the absence of Zds1/Zds2. By genetically manipulating the nucleocytoplasmic distribution of Cdc55, we showed that Cdc55 promotes mitotic entry when in the cytoplasm. On the other hand, nuclear Cdc55 prevents mitotic exit. Our analysis defines the long-sought molecular function for the zillion different screens family proteins and reveals the importance of the regulation of PP2A localization for proper mitotic progression.
Introduction
PP2A (protein phosphatase 2A) is a large family of heterotrimeric phosphatases that account for the majority of serine/threonine phosphatase activity and has important roles in mitotic progression in eukaryotic cells (Shi, 2009). The PP2A complex consists of a structural A subunit, a catalytic C subunit, and a regulatory B subunit, which binds to the AC heterodimer. These B subunits regulate both the substrate specificity and localization of the PP2A complexes.
Two B subunits, Cdc55 (B) and Rts1 (B’), have been identified in budding yeast. Cdc55 and Rts1 bind to the core PP2A subunits in a mutually exclusive manner, and cdc55Δ and rts1Δ exhibit distinct phenotypes, suggesting that they control different functions of PP2A (Shu et al., 1997; Zhao et al., 1997; Jiang, 2006). Cdc55 is important for mitosis, stress response, and polarized growth (Healy et al., 1991; Lin and Arndt, 1995; Minshull et al., 1996; Evans and Stark, 1997; Wang and Burke, 1997; Yang et al., 2000; Jiang, 2006; Queralt et al., 2006; Wang and Ng, 2006; Yellman and Burke, 2006; Chiroli et al., 2007). Cdc55 localizes to various sites, including the bud cortex, the bud neck, the vacuolar membrane, and in the nucleus, and recruits other PP2A subunits (Gentry and Hallberg, 2002). Furthermore, the dephosphorylation of Cdc55–PP2A substrates are cell cycle regulated (Queralt et al., 2006; Pal et al., 2008; Wicky et al., 2011). Thus, it is important to understand the spatiotemporal regulation of Cdc55. However, little is known about the mechanisms that control Cdc55 localization and/or activity.
Recent studies defined Zds1 (zillion different screens 1) and Zds2 proteins as regulators of Cdc55 (Yasutis et al., 2010; Wicky et al., 2011). Zds1 and Zds2 are paralogues and are widely conserved in fungi, including the fission yeast Schizosaccharomyces pombe (Yakura et al., 2006). As implied by their names, Zds1 and Zds2 have been identified as multicopy suppressors of mutants involved in divergent cellular processes, including cell cycle, transcription, and translation, cell polarity, and stress response (Bi and Pringle, 1996; Yu et al., 1996; Mizunuma et al., 1998; Schwer et al., 1998; Roy and Runge, 1999, 2000; Griffioen et al., 2001, 2003; Sekiya-Kawasaki et al., 2002; Hsu et al., 2004; Estruch et al., 2005; Zanelli and Valentini, 2005; Yokoyama et al., 2006). However, the molecular mechanism by which Zds1/Zds2 regulate such divergent processes remains a long-standing mystery.
Accumulating evidences suggest that Zds1/Zds2 directly regulate Cdc55–PP2A function. In large scale proteomics studies, both Zds1 and Zds2 were affinity purified with Cdc55–PP2A (Gavin et al., 2002; Ho et al., 2002; Krogan et al., 2006; Collins et al., 2007). A more recent study revealed that the Zds2 protein directly binds to Cdc55 in vitro via a highly conserved C-terminal region (Yasutis et al., 2010). Furthermore, Zds1 was shown to form a tight stoichiometric complex with Cdc55-containing PP2A (Queralt and Uhlmann, 2008; Wicky et al., 2011). Most importantly, association of Zds1 to PP2A is exclusively mediated via Cdc55 (Wicky et al., 2011). Thus, Zds1/2 is an attractive candidate that specifically controls Cdc55–PP2A and not Rts1–PP2A.
The best-characterized functions of Cdc55 and Zds1/Zds2 are their roles in mitotic entry (Wang and Burke, 1997; McMillan et al., 1999b; Yang et al., 2000; Pal et al., 2008; Yasutis et al., 2010; Wicky et al., 2011). Mitotic entry is driven by activation of the Cdk1 (Nurse, 1975). In budding yeast, Cdc28 (Cdk1) activation is prevented by the Swe1 kinase (Wee1 homologue), which phosphorylates Cdc28 on tyrosine 19 (Tyr-19; Booher et al., 1993; Lew and Reed, 1993), whereas Mih1 phosphatase (Cdc25 homologue) removes this inhibitory phosphorylation to promote mitotic entry (Russell et al., 1989). Because polarized growth and cell cycle are tightly coupled in budding yeast and activation of mitotic Cdc28 depolarizes cell polarity, mutants defective in mitotic entry often lead to prolonged apical bud growth (Lew and Reed, 1993). Both Cdc55 and Zds1/Zds2 are important for mitotic entry, as the cdc55 deletion or zds1Δ zds2Δ double mutant leads to abnormally elongated cell morphology as a consequence of prolonged G2 delay (Healy et al., 1991; Bi and Pringle, 1996; Yu et al., 1996). In these mutants, Swe1 is stabilized (Yang et al., 2000), and Mih1 is hyperphosphorylated (Pal et al., 2008; Wicky et al., 2011). Furthermore, the hyperelongated morphology of cdc55Δ and zds1Δ zds2Δ is rescued either by deletion of SWE1 (McMillan et al., 1999a; Yang et al., 2000), by overexpression of MIH1 (McMillan et al., 1999a), or by introduction of an unphosphorylatable CDC28Y19F mutation (Wang and Burke, 1997; McMillan et al., 1999a; Wicky et al., 2011). Thus, Zds1/Zds2 are thought to function positively with Cdc55–PP2A to promote mitotic entry (Yasutis et al., 2010; Wicky et al., 2011).
Although Zds1/Zds2 forms a tight complex with Cdc55 and function together with Cdc55–PP2A for mitotic entry, it is not always promoting Cdc55–PP2A functions. For example, Cdc55 is required for spindle assembly checkpoint (Minshull et al., 1996; Wang and Burke, 1997; Yellman and Burke, 2006), but Zds1/Zds2 are not required for the spindle assembly checkpoint (Wang and Burke, 1997). Furthermore, a recent paper demonstrated that Zds1/Zds2 is an inhibitor of Cdc55–PP2A during mitotic exit (Queralt and Uhlmann, 2008).
Cdc55–PP2A prevents mitotic exit by inhibition of a Cdc14 phosphatase (Queralt et al., 2006; Wang and Ng, 2006; Yellman and Burke, 2006). Cdc14 promotes mitotic exit by counteracting with Cdc28-dependent phosphorylation (Visintin et al., 1998). Cdc14 is kept inactive in the nucleolus by its inhibitor Net1/Cfi1 (Straight et al., 1999; Visintin et al., 1999). Phosphorylation of Net1/Cfi1 by Cdk1 results in Cdc14 release and activation during anaphase (Azzam et al., 2004). Cdc55 antagonizes this Net1/Cfi1 phosphorylation and prevents Cdc14 release, and Cdc55–PP2A activity toward Net1/Cfi1 is inhibited at the onset of anaphase (Queralt et al., 2006). Surprisingly, Zds1/Zds2 play opposite roles in mitotic exit. In contrast to Cdc55, Zds1/Zds2 are required for Net1/Cfi1 phosphorylation and optimal Cdc14 release from the nucleolus, and zds1Δ zds2Δ shows a severe defect in mitotic exit (Queralt and Uhlmann, 2008). Because overexpression of Zds1 partially inhibits Cdc55–PP2A catalytic activity toward Net1 and because the mitotic exit defect of zds1Δ zds2Δ is fully rescued by deletion of CDC55, it is proposed that Zds1/Zds2 are inhibitors of Cdc55–PP2A activity (Queralt and Uhlmann, 2008).
Here, we propose that Zds1/Zds2 primarily control Cdc55 localization. Cortical and cytoplasmic localization of Cdc55 requires Zds1/Zds2 because Cdc55 accumulates in the nucleus in the absence of Zds1/Zds2. By genetically manipulating nucleocytoplasmic distribution of Cdc55, we show that Zds1/Zds2 act as positive regulators for cytoplasmic Cdc55–PP2A but as negative regulators for nuclear Cdc55–PP2A functions. Our analysis reveals the importance of the regulation of PP2A localization for proper mitotic progression.
Results and discussion
Zds1 and Zds2 are required for cytoplasmic localization of Cdc55
We first examined subcellular localization of Zds1, Zds2, and Cdc55. To ensure that the proteins were expressed at the native level, we added a GFP sequence to the genes encoding Cdc55, Zds1, and Zds2 at their endogenous locus. These GFP fusion constructs were fully functional.
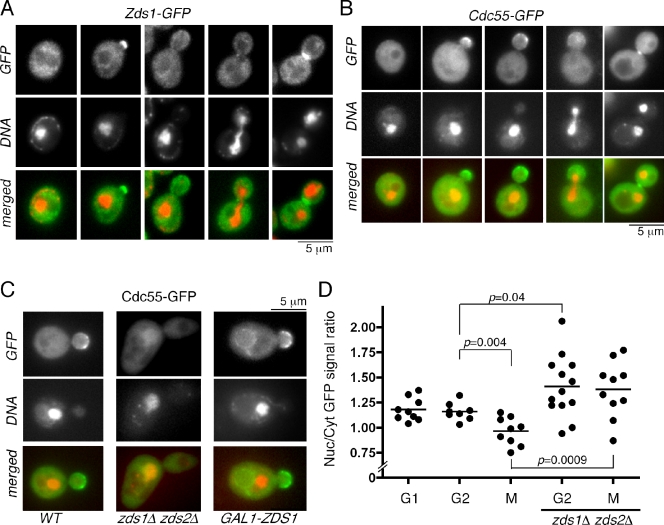
As it has been previously reported with exogenously expressed GST-Zds1, GST-Zds2 (Bi and Pringle, 1996), and Zds2-i-9×myc (Yasutis et al., 2010), both Zds1-GFP and Zds2-GFP localized to the bud cortex of small to medium budded cells (Fig. 1 A and Fig. S1). We also observed bud neck localization of Zds1-GFP and Zds2-GFP in late mitotic cells (Fig. 1 A and Fig. S1), which is consistent with the previous observation of GST-Zds1 localization (Bi and Pringle, 1996). Similarly, Cdc55-GFP localized to the bud cortex of small to medium budded cells and to the bud neck in late mitotic cells that is consistent with a previous study using GFP-Cdc55 (Fig. 1 B; Gentry and Hallberg, 2002). Zds1, Zds2, and Cdc55 also exist in the cytoplasm and are excluded from the vacuole (Fig. 1, A and B).
Figure 1.
Localization of Zds1 and Cdc55. (A) Cell cycle localization of endogenous Zds1-GFP. Cells were fixed with formaldehyde and stained with DAPI. (B) Cell cycle localization of endogenous Cdc55-GFP. In live cells, DNA was stained with Hoechst 33258. (C) Nuclear localization of Cdc55-GFP is affected in zds1Δ zds2Δ cells and in the cells overexpressing Zds1 (GAL1-ZDS1). (D) Nuclear accumulation of Cdc55-GFP is prevented by Zds1 and Zds2. The ratio of mean fluorescence intensity of nuclear (Nuc) and cytoplasmic (Cyt) Cdc55-GFP in each individual cell was plotted. Horizontal lines indicate means. n = 8–13 for each category. G1, unbudded cells; G2, budded with a single nucleus; M, large budded cells with dividing or divided nuclei. P-value was calculated by Student’s unpaired t test.
Interestingly, Zds1 and Zds2 were excluded from the nucleus, as judged by DAPI staining of the nucleus throughout the cell cycle (Fig. 1 A and Fig. S1). Nuclear exclusion of Zds1-GFP was also observed when it was highly overexpressed under a strong GAL1 promoter. Bi and Pringle (1996) also reported that GST-Zds1 and GST-Zds2 are exclusively cytoplasmic and that nuclear localization was hardly detected. These observations suggest that Zds1 and Zds2 most likely function at the bud cortex and in the cytoplasm but not in the nucleus. In contrast, Cdc55-GFP was localized also in the nucleus (Fig. 1 B). Nuclear localization of Cdc55-GFP was observed throughout the cell cycle, which is consistent with the previous study using GFP-Cdc55 (Gentry and Hallberg, 2002) or 3HA-Cdc55 (Queralt et al., 2006) and with the fact that several Cdc55–PP2A targets, such as Esp1 and Net1, are localized in the nucleus or the nucleolus (Queralt et al., 2006; Clift et al., 2009).
To gain further insight into Zds1 and Cdc55 localization, we examined the interdependence of their localization. Localization of Zds1-GFP to the bud cortex and to the bud neck was severely impaired in cdc55Δ but still remained excluded from the nucleus (Fig. S2). We also examined the localization of Zds1ΔC800-GFP, which lacks the C-terminal Cdc55-binding domain (Yasutis et al., 2010). Zds1ΔC800-GFP localization to the bud cortex and at the bud neck was also defective but still excluded from nucleus (Fig. S2). Similarly, Cdc55-GFP failed to localize to the bud cortex and at the bud neck in the zds1Δ zds2Δ cells (Fig. 1 C). Thus, cortical and bud neck localization of Zds1 and Cdc55 is interdependent.
Interestingly, Cdc55-GFP accumulated more in the nucleus in the zds1Δ zds2Δ cells (Fig. 1 C). In contrast, overexpression of ZDS1 from a strong GAL1 promoter resulted in enhancement of cortical, cytoplasmic, and vacuolar membrane localization of Cdc55-GFP and a reduction of the GFP signal from the nucleus (Fig. 1 C).
Quantification of mean GFP fluorescence intensity in the nucleus and in the cytoplasm further confirmed that Zds1/Zds2 affect the nuclear to cytoplasmic ratio of Cdc55-GFP. In wild-type cells, Cdc55-GFP was more concentrated in the nucleus for cells both in G1 and G2 than cells in mitosis (Fig. 1 D). Reduction of nuclear Cdc55-GFP signal in mitosis was dependent on Zds1/Zds2 (Fig. 1 D). Nuclear Cdc55-GFP signal was significantly increased not only in mitotic cells but also in G2 cells for the zds1Δ zds2Δ strain (Fig. 1 D), suggesting that Zds1/Zds2 prevent nuclear accumulation of Cdc55 throughout the cell cycle. These observations prompted us to test whether Zds1/Zds2 promote mitotic progression by preventing nuclear accumulation of Cdc55.
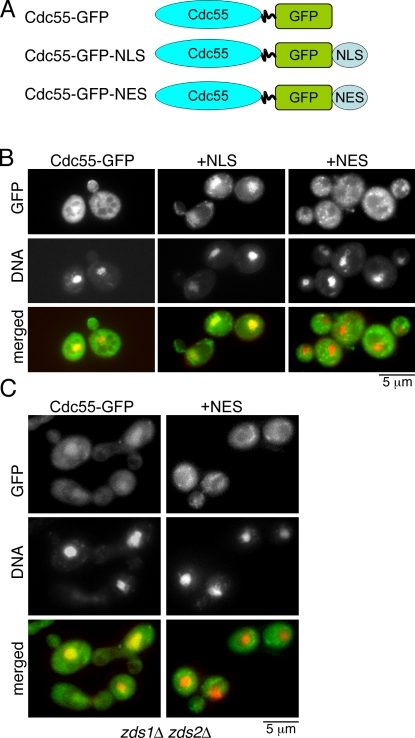
CDC55 mutants specifically targeted to the cytoplasm or the nucleus
To test whether the nucleocytoplasmic distribution of Cdc55 is important for proper mitotic progression, we first generated two novel mutants of CDC55 (Fig. 2 A). By adding a strong nuclear export signal (NES; AAALALKLAGLNI; Hodel et al., 2006) to Cdc55-GFP, we were able to make a cdc55-NES mutant, which is constitutively cytoplasmic (Fig. 2 B) even in the absence of both Zds1/Zds2 (Fig. 2 C). We also made a cdc55-NLS mutant by the addition of a strong NLS of the SV40 antigen (AAAPKKKRKVG; Hodel et al., 2006), which is constitutively nuclear (Fig. 2 B). With these unique cdc55 mutants, we are now able to test our hypothesis that nucleocytoplasmic distribution of Cdc55 under the control of Zds1 is important for mitotic progression.
Figure 2.
Generation of nuclear and cytoplasmic Cdc55 mutants. (A) Schematic representation of Cdc55 mutants. (B) Localization of Cdc55-NLS and Cdc55-NES in living cells. (C) Localization of Cdc55 and Cdc55-NES in zds1Δ zds2Δ cells. For all images in B and C, the DNA was stained with Hoechst 33258.
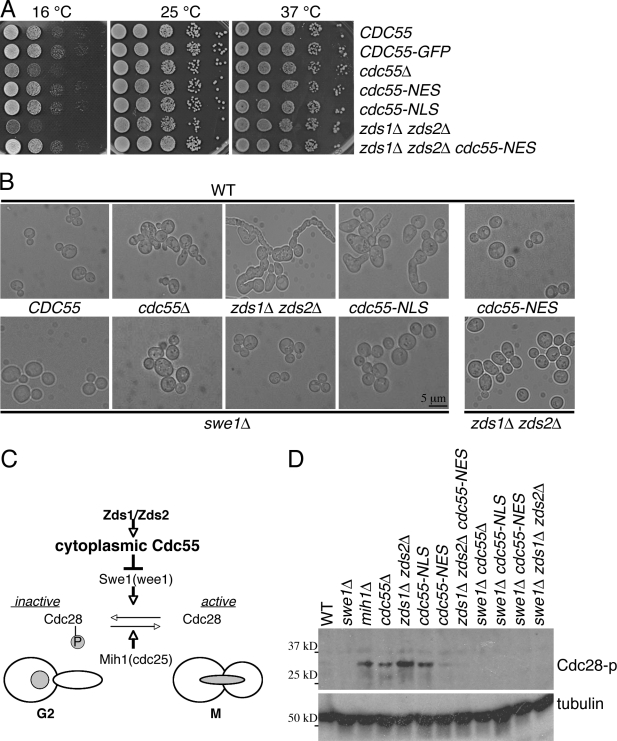
Cytoplasmic Cdc55 and Zds1/Zds2 promote mitotic entry
In our strain background (BY4741), all the cdc55Δ, zds1Δ, zds2Δ, or zds1Δ zds2Δ strains were viable. However, cdc55Δ and zds1Δ zds2Δ strains exhibited severe growth defects at a lowered temperature (Fig. 3 A) as it has been previously described in the other genetic background (Bi and Pringle, 1996; Minshull et al., 1996). The cold-sensitive growth of these mutants likely reflects defective mitotic entry, as the cold-sensitive growth defect of cdc55 is rescued by a CDC28Y19F mutation (Wang and Burke, 1997). Consistent with their positive roles in mitotic entry, both cdc55Δ and zds1Δ zds2Δ exhibited abnormally elongated bud morphology at all the temperatures tested (Fig. 3 B and Fig. S3 B). In agreement with a similar domain analysis on Zds2 (Yasutis et al., 2010), we also found that a conserved Cdc55-binding region of Zds1 was important for Zds1 functions. In a zds2Δ background, Zds1 mutants lacking the Cdc55-binding domain (zds1ΔC800 and zds1ΔC400) exhibited growth defects at a low temperature and a hyperelongated morphology similar to zds1Δ (Fig. S3 C). Furthermore, overexpression of the Cdc55-binding domain of Zds1 (aa 801–913) was sufficient to rescue the cold sensitivity and elongated morphology of zds1Δ zds2Δ cells (Fig. S3 D). Thus, Zds1 function in mitotic entry is likely mediated via Cdc55 binding.
Figure 3.
Zds1/Zds2 and cytoplasmic Cdc55 promote mitotic entry. (A) Serial dilutions of indicated strains spotted on YPD (yeast peptone dextrose) at different temperatures. (B) Representative images of cells with the indicated genotypes in the presence (top row) or absence (bottom row) of SWE1 except for cdc55-NES zds1Δ zds2Δ. Cells were grown in YPD media at 24°C. (C) Our model of mitotic entry regulation by Zds1/Zds2 and cytoplasmic Cdc55. (D) Cytoplasmic Cdc55 is required for dephosphorylation of Cdc28-Y19. Cells were treated with 15 µg/ml nocodazole for 3 h to prevent mitotic progression before cell lysate extraction for Western blotting. Antiphospho–Cdc2-Y15 and antitubulin (loading control) antibodies were used for detection. P, phosphorylation. WT, wild type.
cdc55-NES mutant cells did not display abnormal morphology. However, cdc55-NLS mutant cells displayed a highly elongated morphology like cdc55Δ or zds1Δ zds2Δ mutants (Fig. 3 B). To confirm that the elongated morphology is caused by a delay in mitotic entry, we deleted SWE1 from these strains. Consistent with the idea that Zds1/Zds2 and Cdc55 promote mitotic entry via prevention of the inhibitory phosphorylation of Cdc28 by Swe1 (Fig. 3 C), deletion of SWE1 was sufficient to suppress the elongated morphology of cdc55Δ, zds1Δ zds2Δ, and cdc55-NLS cells (Fig. 3 B).
Next, we confirmed that the elongated morphology of cdc55Δ and zds1Δ zds2Δ mutants was indeed caused by abnormal phosphorylation of Cdc28 at Tyr-19 (Minshull et al., 1996; Yang et al., 2000; Pal et al., 2008). We synchronized the cells in G2/M by nocodazole (a microtubule-destabilizing drug) treatment for 3 h and monitored phosphorylation status of Cdc28 by Western blotting using a phosphospecific antibody specific to Cdc28-Y19. As expected, the levels of Cdc28-Y19 phosphorylation were high in the cells lacking the Cdc28 phosphatase mih1Δ and were absent in the swe1Δ cells (Fig. 3 D). Mutants that showed the elongated morphology (cdc55Δ, zds1Δ zds2Δ, and cdc55-NLS) have significantly elevated levels of Cdc28-Y19 phosphorylation, suggesting a prolonged delay in G2, and this phosphorylation was fully eliminated by deleting SWE1 in these strains (Fig. 3 D). These morphological and biochemical data are consistent with the idea that both Cdc55 and Zds1/Zds2 function together to inactivate Swe1 for mitotic entry.
The observation that the cdc55-NLS (nuclear), but not cdc55-NES (cytoplasmic), mutant was defective in mitotic entry suggests that the cytoplasmic localization of Cdc55 is important for mitotic entry. To test this hypothesis, we examined whether cdc55-NES can bypass the requirement of Zds1/Zds2 not only for cytoplasmic localization of Cdc55 (Fig. 2 C) but also for mitotic entry. Indeed, both the abnormally elongated morphology (Fig. 3 B) and elevated phosphorylation of Cdc28-Y19 in zds1Δ zds2Δ cells (Fig. 3 D) were almost fully rescued by the cdc55-NES mutation. Furthermore, the cold-sensitive growth defect of zds1Δ zds2Δ was also rescued by the cdc55-NES (Fig. 3 A). Thus, the cytoplasmic localization of Cdc55 mediated by Zds1/Zds2 is required and sufficient for normal mitotic entry. The finding that the cdc55-NES mutation bypassed the requirement of Zds1/Zds2 for mitotic entry suggests that the most important function of Zds1/Zds2 in mitotic entry is to promote Cdc55 export from the nucleus or to maintain Cdc55 in the cytoplasm.
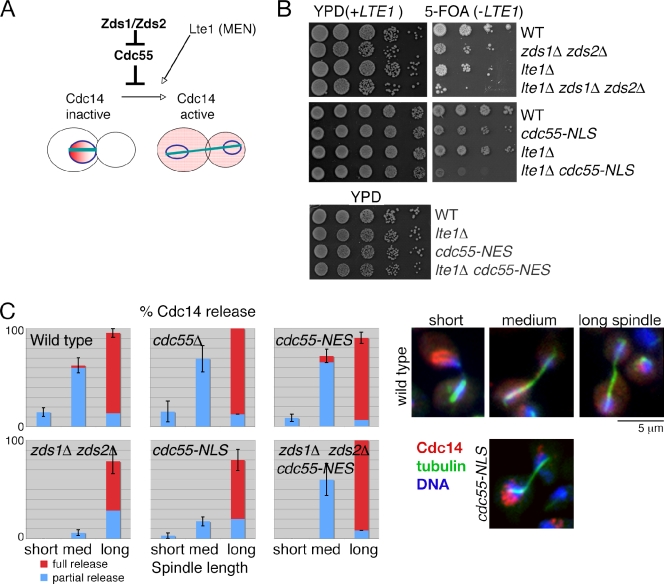
Nuclear Cdc55 interferes with mitotic exit
It is known that Cdc55–PP2A is an inhibitor of mitotic exit (Queralt et al., 2006; Wang and Ng, 2006; Yellman and Burke, 2006). Cdc55 inhibits Cdc14 release from the nucleolus, a key step for mitotic exit, by preventing Net1/Cfi1 phosphorylation. In contrast, Zds1/Zds2 were recently shown to be activators of mitotic exit (Queralt and Uhlmann, 2008). Because Zds1/Zds2 and Cdc55 physically interact and the mitotic exit defect of the zds1Δ zds2Δ cells was suppressed by deletion of CDC55, it has been proposed that Zds1/Zds2 are inhibitors of Cdc55–PP2A activity (Fig. 4 A).
Figure 4.
Nuclear Cdc55 prevents mitotic exit by inhibiting Cdc14 release from the nucleolus. (A) Model of Cdc14 release controlled by Cdc55 and Zds1/Zds2 proteins. (B) Nuclear Cdc55 is toxic to lte1Δ mutant cells. Serial dilutions of the indicated strains containing a complementing LTE1 plasmid (with a URA3 marker) were grown on YPD plates or plates containing 5-fluoroorotic acid (5-FOA) to counter the select LTE1 plasmid at 30°C for 3 d (top). cdc55-NES and lte1Δ are not synthetically lethal. Serial dilutions of the indicated strains were spotted on YPD at 24°C (bottom). (C) Quantification of Cdc14 release during anaphase. Mitotic cells were categorized into short spindle (0–3 µm), medium (med) spindle (3–7 µm), and long spindle (>7 µm). In each category, Cdc14 release status was classified as partial release or full release. Error bars are SEM. n = 133 (wild type), 33 (cdc55), 122 (cdc55-NES), 123 (cdc55-NLS), 107 (zds1 zds2), and 65 (zds1 zds2 cdc55-NES). Examples of wild-type Cdc14 localization during anaphase are on the top right, and an example of a midanaphase cdc55-NLS cell is shown on the bottom right. Cdc14 and tubulin were visualized by indirect immunofluorescence microscopy using the anti-Cdc14 antibody and α-tubulin antibody. DNA was stained with DAPI. MEN, mitotic exit network.
To examine the role of Cdc55 and Zds1/Zds2 proteins in mitotic exit, we first tested for genetic interactions using LTE1 deletion strains. Lte1 is a nonessential component of the mitotic exit network and shows synthetic lethality with genes involved in the early anaphase release of Cdc14 (FEAR) pathway (Stegmeier et al., 2002). Consistent with the requirement of Zds1/Zds2 in the FEAR pathway, we found a synthetic lethality between lte1Δ and zds1Δ zds2Δ (Fig. 4 B). We hypothesized that this synthetic lethality is derived from the nuclear accumulation of Cdc55. Consistent with our hypothesis, cdc55-NLS showed synthetic lethality with lte1Δ (Fig. 4 B). In contrast, cdc55-NES showed no synthetic growth defects with lte1Δ (Fig. 4 B).
To further confirm that nuclear Cdc55 is preventing the FEAR pathway, we analyzed the timing of Cdc14 release from the nucleolus using spindle length as an internal marker for mitotic progression. Wild-type, cdc55Δ, and cdc55-NES cells partially released Cdc14 in early anaphase (spindle length of 3–7 µm) and fully released Cdc14 in late mitosis (spindle length >7 µm; Fig. 4 C). In contrast, Cdc14 release in early anaphase was significantly impaired in zds1Δ zds2Δ and cdc55-NLS cells (Fig. 4 C). Furthermore, Cdc14 release was partially impaired in the later stages of mitosis in zds1Δ zds2Δ and cdc55-NLS cells (Fig. 4 C). These results suggest that nuclear Cdc55 interferes with mitotic exit by inhibiting the release of Cdc14 from the nucleolus. In support of this hypothesis, artificial exclusion of Cdc55 from the nucleus by the cdc55-NES mutation fully rescued Cdc14 release defects of the zds1Δ zds2Δ mutant (Fig. 4 C). Thus, exclusion of Cdc55 from the nucleus is the key function of Zds1/Zds2 to promote mitotic exit.
Zds1/Zds2 promote mitotic entry via Cdc55 function in the cytoplasm
In this study, we show that Zds1/Zds2 proteins promote cytoplasmic functions of Cdc55–PP2A (Fig. 5). In the absence of Zds1/Zds2, Cdc55 accumulates in the nucleus, resulting in defective mitotic entry. Reduction of cytoplasmic Cdc55 is likely the reason for the G2 delay because the cdc55-NLS mutant, whose gene product is dominantly nuclear, phenocopies zds1Δ zds2Δ. The G2 delay is not caused by the nuclear accumulation of Cdc55 because the elongated bud morphology of cdc55-NLS was rescued by an extra copy of CDC55 (Fig. S2). In contrast, the cdc55-NES mutant, whose gene product is dominantly cytoplasmic, is fully competent in mitotic entry and can even rescue the mitotic entry defects of zds1Δ zds2Δ cells. Our data suggest that the primary function of Zds1/Zds2 during the cell cycle is to keep Cdc55–PP2A in the cytoplasm because not only the mitotic entry defect but also the cold-sensitive growth defect of the zds1Δ zds2Δ cells was rescued by the cdc55-NES. This bypass effect also suggests that Zds1/Zds2 is not required for the catalytic activity or substrate specificity of Cdc55–PP2A in the cytoplasm.
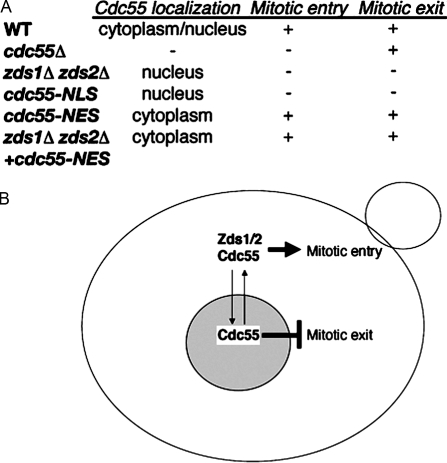
Figure 5.
Summary and model of Zds1/Zds2–Cdc55–PP2A regulation of mitosis. (A) Summary table of Cdc55 localization and mitotic entry and exit defects. (B) A model of the regulation of mitosis by Cdc55 and Zds1/Zds2 proteins. When bound to Zds1/Zds2, Cdc55 functions in the cytoplasm to promote mitotic entry by activating the Cdk. Exclusion of Cdc55 from the nucleus by Zds1/Zds2 proteins is also required for mitotic exit by preventing Cdc14 release. In the absence of Zds1/Zds2, Cdc55 remains in the nucleus, leading to a delay in mitotic entry and mitotic exit. See Results and discussion for more details. WT, wild type.
Our genetic data suggest that the critical target of Cdc55 in mitotic entry is Swe1 because mitotic entry defects of cdc55Δ, zds1Δ zds2Δ, and cdc55-NLS are almost fully rescued by deletion of SWE1. Dephosphorylation of Mih1 is regulated by Cdc55 (Pal et al., 2008; Wicky et al., 2011). Therefore, Mih1 is also a target of Zds1/Zds2–Cdc55–PP2A. However, mih1Δ cells have a relatively milder defect in mitotic entry (Russell et al., 1989; Pal et al., 2008) compared with the cdc55Δ or the zds1Δ zds2Δ, suggesting that Mih1 is not the only target. It is important to mention that both Swe1 and Mih1 are regulated by nucleocytoplasmic transport, and a swe1 mutant defective in nuclear export is a more potent inhibitor of mitotic entry (Keaton et al., 2008). Thus, we speculate that Swe1 inactivation by Zds1/Zds2–Cdc55–PP2A takes place in the cytoplasm, which is also consistent with the fact that Swe1-inactivating kinases are also in the cytoplasm (McMillan et al., 1999a; Shulewitz et al., 1999; Asano et al., 2005).
The role and significance of the bud tip and the bud neck localization of Zds1/Zds2 and Cdc55 remain an area of interest for future research. Zds1/Zds2 interact with cell polarity proteins of the Cdc42- and the Rho1-signaling pathways both physically and genetically (Bi and Pringle, 1996; Drees et al., 2001; Sekiya-Kawasaki et al., 2002), we speculate that Zds1/Zds2 also have direct roles in cell polarity beyond controlling Cdk1 activity. Alternatively, cell polarity factors might influence cell cycle progression via Zds1/Zds2–Cdc55 regulation.
Zds1/Zds2 promote mitotic exit by nuclear exclusion of Cdc55
It was proposed that Zds1/Zds2, together with the separase Esp1, inhibit Cdc55–PP2A catalytic activity to promote Cdc14 release in anaphase (Queralt and Uhlmann, 2008). Given the fact that Esp1, Cdc55, and its substrate Net1 are all in the nucleus and/or nucleolus, the Zds1/Zds2-mediated inhibition of Cdc55–PP2A must occur within the nucleus. Our genetic data are generally consistent with Queralt and Uhlmann (2008), and we do not exclude the possibility that Zds1/Zds2 directly inhibit Cdc55–PP2A activity specific to Net1, but the following evidence supports our hypothesis that Zds1/Zds2 promotes mitotic exit primarily by excluding Cdc55 from the nucleus (Fig. 5).
First, the majority of Zds1/Zds2 is in the cytoplasm. Given that Zds1 forms a stoichiometric complex with Cdc55–PP2A, it is unlikely that a minor fraction of Zds1/Zds2 in the nucleus is sufficient for inhibiting Cdc55–PP2A in the nucleus. Second, nuclear-accumulated Cdc55-NLS dominantly inhibits mitotic exit even in the presence of Zds1/Zds2, suggesting that nuclear Cdc55–PP2A activity is not effectively inhibited by Zds1/Zds2. Third, the nuclear Cdc55 signal is significantly reduced during mitosis in a Zds1/Zds2-dependent manner. It is important to mention that reduction of Cdc55 intensity from the nucleus was underestimated in our assay because it was impossible to exclude contaminating strong cytoplasmic signals from other focal planes with our standard fluorescent microscope. Our genetic data are also consistent with our hypothesis that nuclear exclusion of Cdc55 is the key function of Zds1/Zds2 because zds1Δ zds2Δ defects in Cdc14 release are fully rescued by the cdc55-NES mutation.
All these lines of evidence suggest that either cytoplasmic Cdc55 or nuclear exclusion of Cdc55 is important for mitotic exit. We favor the latter hypothesis because deletion of CDC55 is sufficient for rescuing the mitotic exit defects of zds1Δ zds2Δ (Queralt and Uhlmann, 2008), and overexpression of CDC55 prevents mitotic exit and is toxic to mitotic exit network mutants (Wang and Ng, 2006).
Evolutional conservation of PP2A regulation
Recent studies revealed that the PP2A–B55 (a counterpart of PP2A–Cdc55) complex is the major regulator of mitotic entry and exit in animal cells (Mochida et al., 2009; Schmitz et al., 2010). In the Xenopus laevis egg and in HeLa cells, PP2A–B55 activity is directly inhibited by Arpp19 and α-endosulfine during mitosis (Gharbi-Ayachi et al., 2010; Mochida et al., 2010). Budding yeast Igo1/Igo2 share sequence homology to Arpp19 and α-endosulfine (Dulubova et al., 2001; Talarek et al., 2010), suggesting a possible conservation of PP2A inhibition during mitosis.
On the other hand, Zds1 is highly conserved in fungi, including pathogenic Candida albicans and Cryptococcus neoformans, but no obvious homologue was found in higher eukaryotes. Considering the fact that a key function of Zds1/Zds2 is to regulate the nucleocytoplasmic distribution of Cdc55–PP2A, it is possible that Zds1/Zds2 has uniquely evolved in fungi, which undergo “closed mitosis,” but is not required in higher eukaryotes, which undergo “open mitosis.” Alternatively, it is also possible that functional counterparts to the zillion different screens protein family exist in higher eukaryotes but are not easily recognizable by their primary sequences, such as the case for Cdk inhibitors.
Materials and methods
Yeast genetics
All yeast strains used in this study were isogenic or congenic to BY4741 (MATa leu2Δ0 his3Δ1 met15Δ0 ura3Δ0, obtained from Thermo Fisher Scientific). Standard yeast genetics was used to generate the strains. Yeast strains are listed in Table S1. PY3295 and SY strains were gifts from D. Pellman (Dana-Farber Cancer Institute, Boston, MA). D. Lew (Duke University, Durham, NC) provided a SWE1 gene knockout plasmid. Gene deletions or modifications were performed with PCR-mediated one-step gene replacement using pFA6a vectors provided by J. Pringle (Stanford University, Stanford, CA; Longtine et al., 1998) and confirmed by PCR. For tagging CDC55, ZDS1, and ZDS2 with GFP, a flexible amino acid linker (GGSGGS) was introduced between the target protein and GFP. For generation of the cdc55-NES and cdc55-NLS mutants, the NES sequence (AAALALKLAGLNI) and SV40 NLS sequence (AAAPKKKRKVG) were directly introduced after the GFP sequence of CDC55-GFP by PCR-mediated one-step gene replacement (Longtine et al., 1998).
Biochemistry
Protein extracts were prepared as previously described (von der Haar, 2007). In brief, cells were treated with the solution containing 0.1 N NaOH, 0.05 M EDTA, 2% SDS, and 2% β-mercaptoethanol and immediately boiled for 10 min. The samples were neutralized with acetic acid, and SDS sample buffer (Boston Bioproducts) was added. Rabbit antiphospho–Cdc2-Y15 antibody (Cell Signaling Technology) and mouse anti-GFP antibody (Millipore) were purchased from the commercial source. Rabbit anti-Rho1 antibody was custom made. Rabbit antitubulin antibody is a gift from B. Goode (Brandeis University, Waltham, MA). HRP-conjugated secondary antibodies were obtained from Millipore, and proteins were detected by an enhanced chemiluminescence system (ECL Plus; GE Healthcare).
Fluorescence microscopy
Fluorescence images were acquired with a fluorescence microscope (Eclipse E600; Nikon) equipped with a charge-coupled device camera (DC350F; Andor) with 100×, NA of 1.45, or 60×, NA 1.4, oil objectives. The images were captured and analyzed with NIS-Elements software (Nikon), and the figures were processed and assembled in Photoshop (Adobe).
For indirect immunofluorescence methods, we fixed the cells with 4% formaldehyde for 10 min at room temperature, and the cell walls were digested with Zymolyase 20T (Zymo Research). The mitotic spindle was immunostained with the YOL1/34 rat monoclonal antibody (AbD Serotec) followed by an FITC-conjugated anti–rat antibody (Jackson ImmunoResearch Laboratories, Inc.). Cdc14 was immunostained with sc-12045 polyclonal antibody (Santa Cruz Biotechnology, Inc.) followed by a CY3-conjugated anti–goat antibody (Rockland Immunochemicals, Inc.). DAPI was used to visualize the nucleus in fixed cells. For live-cell imaging, all strains were grown in synthetic media at room temperature. We used Hoechst 33258 (AnaSpec) at a 2-µM concentration in PBS to stain the nucleus. For quantifying the nucleocytoplasmic ratio of Cdc55-GFP, mean nuclear fluorescence intensity and mean cytoplasmic fluorescence intensity were calculated with NIS-Elements software.
Online supplemental material
Fig. S1 shows that Zds2, like Zds1, is excluded from the nucleus. Fig. S2 shows that cortical and bud neck localization of Zds1 and Cdc55 are interdependent. Fig. S3 shows that the conserved Cdc55-binding region of Zds1 is important for Zds1 function in mitotic entry. Table S1 contains the list of yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101134/DC1.
Acknowledgments
We thank David Pellman for the discussion and support. We thank David Pellman, Daniel Lew, John Pringle, and Bruce Goode for the gift of reagents and Erin Jonasson for her critical reading of the manuscript and for discussion.
This paper was supported by a Massachusetts Life Sciences Center grant and by a start-up fund of Brandeis University.
Footnotes
Abbreviations used in this paper:
- FEAR
- early anaphase release of Cdc14
- NES
- nuclear export signal
References
- Asano S., Park J.E., Sakchaisri K., Yu L.R., Song S., Supavilai P., Veenstra T.D., Lee K.S. 2005. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 24:2194–2204 10.1038/sj.emboj.7600683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., Deshaies R.J. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 305:516–519 10.1126/science.1099402 [DOI] [PubMed] [Google Scholar]
- Bi E., Pringle J.R. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5264–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R.N., Deshaies R.J., Kirschner M.W. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiroli E., Rossio V., Lucchini G., Piatti S. 2007. The budding yeast PP2ACdc55 protein phosphatase prevents the onset of anaphase in response to morphogenetic defects. J. Cell Biol. 177:599–611 10.1083/jcb.200609088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D., Bizzari F., Marston A.L. 2009. Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev. 23:766–780 10.1101/gad.507509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.R., Kemmeren P., Zhao X.C., Greenblatt J.F., Spencer F., Holstege F.C., Weissman J.S., Krogan N.J. 2007. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics. 6:439–450 [DOI] [PubMed] [Google Scholar]
- Drees B.L., Sundin B., Brazeau E., Caviston J.P., Chen G.C., Guo W., Kozminski K.G., Lau M.W., Moskow J.J., Tong A., et al. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549–571 10.1083/jcb.200104057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Horiuchi A., Snyder G.L., Girault J.A., Czernik A.J., Shao L., Ramabhadran R., Greengard P., Nairn A.C. 2001. ARPP-16/ARPP-19: a highly conserved family of cAMP-regulated phosphoproteins. J. Neurochem. 77:229–238 10.1046/j.1471-4159.2001.t01-1-00191.x [DOI] [PubMed] [Google Scholar]
- Estruch F., Hodge C.A., Rodríguez-Navarro S., Cole C.N. 2005. Physical and genetic interactions link the yeast protein Zds1p with mRNA nuclear export. J. Biol. Chem. 280:9691–9697 10.1074/jbc.M413025200 [DOI] [PubMed] [Google Scholar]
- Evans D.R., Stark M.J. 1997. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics. 145:227–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- Gentry M.S., Hallberg R.L. 2002. Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol. Biol. Cell. 13:3477–3492 10.1091/mbc.02-05-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi-Ayachi A., Labbé J.C., Burgess A., Vigneron S., Strub J.M., Brioudes E., Van-Dorsselaer A., Castro A., Lorca T. 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science. 330:1673–1677 10.1126/science.1197048 [DOI] [PubMed] [Google Scholar]
- Griffioen G., Branduardi P., Ballarini A., Anghileri P., Norbeck J., Baroni M.D., Ruis H. 2001. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell. Biol. 21:511–523 10.1128/MCB.21.2.511-523.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen G., Swinnen S., Thevelein J.M. 2003. Feedback inhibition on cell wall integrity signaling by Zds1 involves Gsk3 phosphorylation of a cAMP-dependent protein kinase regulatory subunit. J. Biol. Chem. 278:23460–23471 10.1074/jbc.M210691200 [DOI] [PubMed] [Google Scholar]
- Healy A.M., Zolnierowicz S., Stapleton A.E., Goebl M., DePaoli-Roach A.A., Pringle J.R. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183 10.1038/415180a [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Harreman M.T., Pulliam K.F., Harben M.E., Holmes J.S., Hodel M.R., Berland K.M., Corbett A.H. 2006. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. J. Biol. Chem. 281:23545–23556 10.1074/jbc.M601718200 [DOI] [PubMed] [Google Scholar]
- Hsu C.L., Chen Y.S., Tsai S.Y., Tu P.J., Wang M.J., Lin J.J. 2004. Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p and Zds2p is involved in telomere replication, telomere maintenance and cell growth control. Nucleic Acids Res. 32:511–521 10.1093/nar/gkh203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. 2006. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:440–449 10.1128/MMBR.00049-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton M.A., Szkotnicki L., Marquitz A.R., Harrison J., Zyla T.R., Lew D.J. 2008. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol. Biol. Cell. 19:4006–4018 10.1091/mbc.E08-03-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440:637–643 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- Lew D.J., Reed S.I. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305–1320 10.1083/jcb.120.6.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.C., Arndt K.T. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- McMillan J.N., Longtine M.S., Sia R.A., Theesfeld C.L., Bardes E.S., Pringle J.R., Lew D.J. 1999a. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 19:6929–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J.N., Sia R.A., Bardes E.S., Lew D.J. 1999b. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol. Cell. Biol. 19:5981–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J., Straight A., Rudner A.D., Dernburg A.F., Belmont A., Murray A.W. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609–1620 10.1016/S0960-9822(02)70784-7 [DOI] [PubMed] [Google Scholar]
- Mizunuma M., Hirata D., Miyahara K., Tsuchiya E., Miyakawa T. 1998. Role of calcineurin and Mpk1 in regulating the onset of mitosis in budding yeast. Nature. 392:303–306 10.1038/32695 [DOI] [PubMed] [Google Scholar]
- Mochida S., Ikeo S., Gannon J., Hunt T. 2009. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 28:2777–2785 10.1038/emboj.2009.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Maslen S.L., Skehel M., Hunt T. 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science. 330:1670–1673 10.1126/science.1195689 [DOI] [PubMed] [Google Scholar]
- Nurse P. 1975. Genetic control of cell size at cell division in yeast. Nature. 256:547–551 10.1038/256547a0 [DOI] [PubMed] [Google Scholar]
- Pal G., Paraz M.T., Kellogg D.R. 2008. Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J. Cell Biol. 180:931–945 10.1083/jcb.200711014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt E., Uhlmann F. 2008. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J. Cell Biol. 182:873–883 10.1083/jcb.200801054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt E., Lehane C., Novak B., Uhlmann F. 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 125:719–732 10.1016/j.cell.2006.03.038 [DOI] [PubMed] [Google Scholar]
- Roy N., Runge K.W. 1999. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma. 108:146–161 10.1007/s004120050364 [DOI] [PubMed] [Google Scholar]
- Roy N., Runge K.W. 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10:111–114 10.1016/S0960-9822(00)00298-0 [DOI] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S.I. 1989. Conservation of mitotic controls in fission and budding yeasts. Cell. 57:295–303 10.1016/0092-8674(89)90967-7 [DOI] [PubMed] [Google Scholar]
- Schmitz M.H., Held M., Janssens V., Hutchins J.R., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A.I., Poser I., et al. 2010. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12:886–893 10.1038/ncb2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Linder P., Shuman S. 1998. Effects of deletion mutations in the yeast Ces1 protein on cell growth and morphology and on high copy suppression of mutations in mRNA capping enzyme and translation initiation factor 4A. Nucleic Acids Res. 26:803–809 10.1093/nar/26.3.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya-Kawasaki M., Abe M., Saka A., Watanabe D., Kono K., Minemura-Asakawa M., Ishihara S., Watanabe T., Ohya Y. 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics. 162:663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2009. Serine/threonine phosphatases: mechanism through structure. Cell. 139:468–484 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Shu Y., Yang H., Hallberg E., Hallberg R. 1997. Molecular genetic analysis of Rts1p, a B’ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz M.J., Inouye C.J., Thorner J. 1999. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7123–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220 10.1016/S0092-8674(02)00618-9 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Shou W., Dowd G.J., Turck C.W., Deshaies R.J., Johnson A.D., Moazed D. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 97:245–256 10.1016/S0092-8674(00)80734-5 [DOI] [PubMed] [Google Scholar]
- Talarek N., Cameroni E., Jaquenoud M., Luo X., Bontron S., Lippman S., Devgan G., Snyder M., Broach J.R., De Virgilio C. 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol. Cell. 38:345–355 10.1016/j.molcel.2010.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., Amon A. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 398:818–823 10.1038/19775 [DOI] [PubMed] [Google Scholar]
- von der Haar T. 2007. Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE. 2:e1078 10.1371/journal.pone.0001078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Burke D.J. 1997. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ng T.Y. 2006. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:80–89 10.1091/mbc.E04-12-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S., Tjandra H., Schieltz D., Yates J., III, Kellogg D.R. 2011. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol. Biol. Cell. 22:20–32 10.1091/mbc.E10-06-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakura M., Ozoe F., Ishida H., Nakagawa T., Tanaka K., Matsuda H., Kawamukai M. 2006. zds1, a novel gene encoding an ortholog of Zds1 and Zds2, controls sexual differentiation, cell wall integrity and cell morphology in fission yeast. Genetics. 172:811–825 10.1534/genetics.105.050906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jiang W., Gentry M., Hallberg R.L. 2000. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol. Cell. Biol. 20:8143–8156 10.1128/MCB.20.21.8143-8156.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutis K., Vignali M., Ryder M., Tameire F., Dighe S.A., Fields S., Kozminski K.G. 2010. Zds2p regulates Swe1p-dependent polarized cell growth in Saccharomyces cerevisiae via a novel Cdc55p interaction domain. Mol. Biol. Cell. 21:4373–4386 10.1091/mbc.E10-04-0326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellman C.M., Burke D.J. 2006. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:658–666 10.1091/mbc.E05-04-0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H., Mizunuma M., Okamoto M., Yamamoto J., Hirata D., Miyakawa T. 2006. Involvement of calcineurin-dependent degradation of Yap1p in Ca2+-induced G2 cell-cycle regulation in Saccharomyces cerevisiae. EMBO Rep. 7:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jiang Y.W., Wellinger R.J., Carlson K., Roberts J.M., Stillman D.J. 1996. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol. 16:5254–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C.F., Valentini S.R. 2005. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 171:1571–1581 10.1534/genetics.105.048082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Boguslawski G., Zitomer R.S., DePaoli-Roach A.A. 1997. Saccharomyces cerevisiae homologs of mammalian B and B’ subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J. Biol. Chem. 272:8256–8262 10.1074/jbc.272.13.8256 [DOI] [PubMed] [Google Scholar]