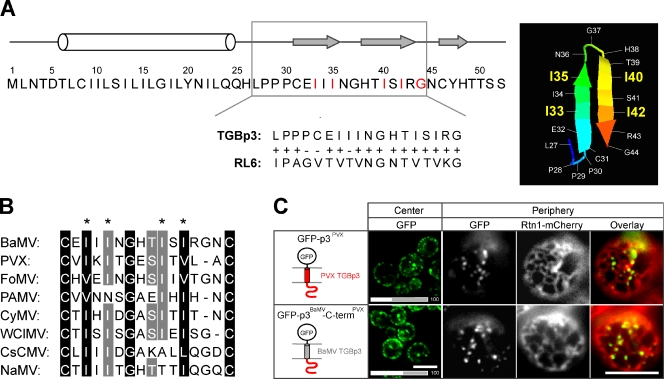
Figure 4.
The critical residues in the sorting signal region of TGBp3 are conserved among the Potexviruses. (A, left) Full-length amino acid sequence and secondary structure of TGBp3 predicted by PSIPRED algorithm (black line, coil; white bar, α helix; gray arrow, β strand; McGuffin et al., 2000). (A, right) The PHYRE algorithm (Kelley and Sternberg, 2009) predicts a hairpin structure in the TGBp3 sorting signal region based on RL6, a 50S ribosomal protein L6 of G. thermodenitrificans (available under GenBank/EMBL/DDBJ accession no. YP_001124252.1). The hairpin region of RL6 aligned with TGBp3 is shown below the TGBp3 sequence. (B) The predicted sorting signals flanked by two cysteine residues of several potexviral TGBp3 were aligned by GeneDoc. The asterisks indicate the four isoleucine residues crucial for BaMV TGBp3 localization. Viral sequences were acquired from the National Center for Biotechnology Information database: BaMV (accession no. NP_042586), PVX (accession no. AAF67821), FoMV (Foxtail mosaic virus, accession no. ABW25057), PAMV (Potato aucuba mosaic virus, accession no. NP_619749), CyMV (Cymbidium mosaic virus, accession no. ABO41880.1), WClMV (White clover mosaic virus, accession no. NP_620718), CsCMV (Cassava common mosaic virus, accession no. NP_042698), and NaMV (Nandina mosaic virus, accession no. AAX19934). (C) Yeast cells expressing GFP-tagged PVX TGBp3 and the hybrid BaMV TGBp3 whose C-terminal region was replaced with that of PVX were imaged by confocal microscopy (center) or coexpressed with Rtn1-mCherry (CWY2754) and imaged by fluorescence microscopy (periphery). The bars on the bottom indicate quantification data. Bars, 5 µm.