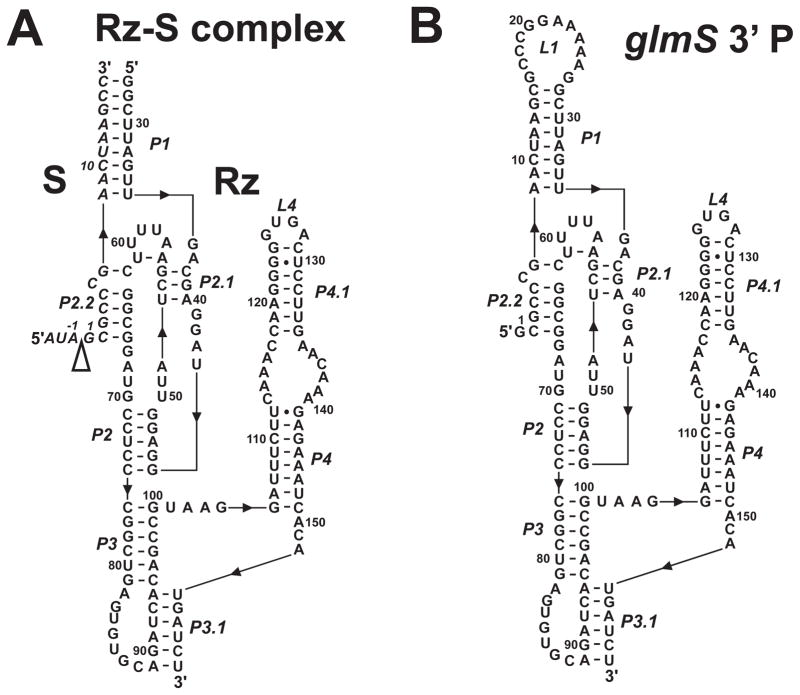
Figure 2.
Secondary structure diagrams of the Bacillus subtilis glmS trans-acting ribozyme-substrate complex (glmS-Rz-S) used in cleavage kinetics experiments (A) and glmS 3′ cleavage product (B) used in hydroxyl radical footprinting. The secondary structures are shown according to existing crystal structures for the glmS ribozyme (12, 14), and numbered according to Winkler and co-workers (6). The 5′ to 3′ direction of the RNA is indicated with black triangles. The cleavage site is indicated with a open triangle.