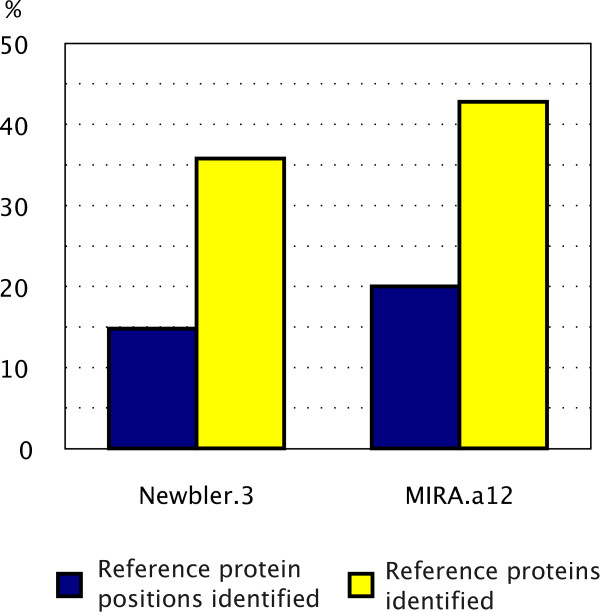
Figure 3.
Comparison of the Newbler.3 and MIRA.a12 assemblers with respect to the numbers of amino acid residues or proteins identified via the est2assembly pipeline. In this E. aurinia dataset, we used the BLASTx similarity to Bombyx mori (cut-off 50 bits) in order to compare performance. MIRA produces an assembly which identifies more of the reference proteome. Further, at this coverage, we do not have a complete coverage of each gene as the proportion of individual amino acids identified is lower (see text for discussion). As this project is a gene-finding one, we choose the MIRA assembly for downstream application.