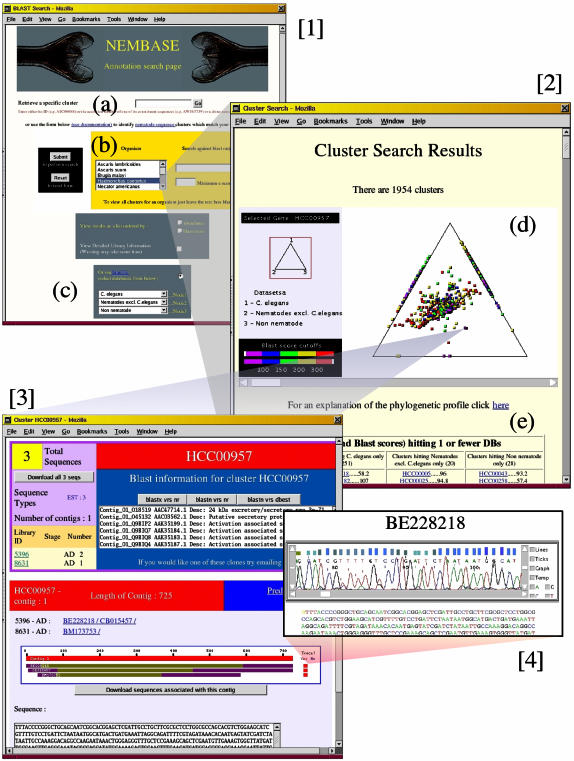
Figure 1.
Screenshots from a typical search strategy on NEMBASE. [1] Annotation search page: on this page users may either retrieve a cluster by entering its ID or the ID of one of its constituent sequences (a) or select a species and enter some text to search for keywords in the BLAST annotation associated with clusters from that species (b). Output may be viewed in terms of relative abundance, BLAST score or using the SimiTri Java tool (c). [2] SimiTri output page: selecting the SimiTri output option creates the embedded Java applet (d); individual clusters are represented by coloured tiles on the graphic. The relative position of the tiles indicates the clusters’ relative similarity to the three selected data sets. Clusters with similarity to only one or no data set are listed below the applet (e). Clicking on a tile whilst holding the control key held down, or selecting a cluster from the list below the applet launches the detailed cluster page [3]. This provides information on the number and source of the constituent sequences, summaries of BLAST annotation and further links to e.g. raw trace chromatograms [4] associated with the sequences.