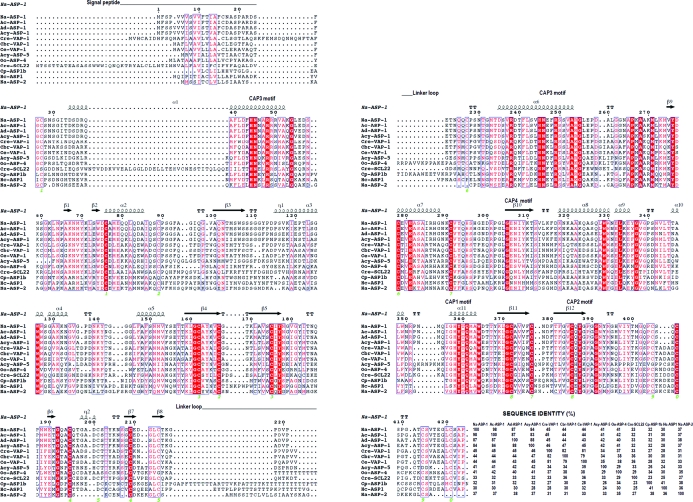
Figure 3.
Comparison of Na-ASP-1 with selected representative two-CAP-domain proteins. Sequence alignment of selected two-CAP-domain proteins reveals the sequence conservation across both domains. The highest variability is in the signal peptide and loop regions. This figure was generated with ESPript. The different secondary-structure elements shown are α-helices as large squiggles labelled α, 310-helices as small squiggles labelled η, β-strands as arrows labelled β and β-turns labelled TT. Identical residues are shown on a red background, conserved residues are shown in red and conserved regions are shown in blue boxes. Na-ASP-2 (AAP41952.1) is included as a representative one-CAP-domain ASP. Aligned proteins are Na-ASP-1, N. americanus ASP-1 (AAD13340.1); Ac-ASP-1, Ancylostoma caninum ASP-1 (AAD318391); Ad-ASP-1, A. duodenale ASP-1 (AAD13339.1); Acy-ASP-1, A. ceylanicum ASP-1 (AAN11402.1); Cre-VAP-1, Caenorhabditis remanei two-domain activation-associated secreted protein (XP_003106746.1); Cbr-VAP-1, C. briggsae VAP-1 (CAP3467.2); Ce-VAP-1, C. elegans VAP-1 (NM_001029382.1); Cre-SCL22, C. remanei two-domain activation-associated secreted protein (XP_003109641.1); Cp-ASP1b, Cooperia punctata two-domain activation-associated secreted protein-like (AAK35199.1); Acy-ASP-5, A. ceylanicum ASP-5 (ABB53347.1); Oo-VAP-4, Ostertagia ostertagi VAP-4 (CA000417); Hc-ASP-1, Haemonchus contortus putative secretory protein precursor (AAC03562). GenBank accession numbers are given in parentheses. The sequence identity of each protein is also listed.