Table 1. Statistics for data collection and model refinement.
Values in parentheses are for the last shell.
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 67.8, b = 74.6, c = 84.7, β = 112.1 |
| Resolution limits (Å) | 30.8–2.2 (2.3–2.2) |
| 〈I/σ(I)〉 | 15.4 (4.7) |
| No. of reflections | 272397 (35542) |
| No. of unique reflections | 37195 (5261) |
| Multiplicity | 7.3 (6.8) |
| Rmerge† (%) | 10.7 (55.7) |
| Completeness (%) | 93.6 (91.4) |
| Rcryst‡ | 0.18 (0.19) |
| Rfree§ | 0.24 (0.26) |
| Correlation coefficients | |
| Fo − Fc | 0.946 |
| Fo − Fc (free) | 0.911 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.020 |
| Bond angles (°) | 1.812 |
| Mean B factors (Å2) | |
| Protein | 16.1 |
| All atoms | 24.5 |
| Model composition | |
| Monomers | 2 |
| Residues | 798 |
| Water molecules | 211 |
R
merge = 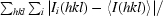
 , where I
i(hkl) and 〈I(hkl)〉 are the intensity of measurement i of I and the mean intensity of the reflection with indices hkl, respectively.
, where I
i(hkl) and 〈I(hkl)〉 are the intensity of measurement i of I and the mean intensity of the reflection with indices hkl, respectively.
R
cryst = 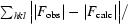
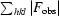 , where F
obs are observed and F
calc are calculated structure-factor amplitudes.
, where F
obs are observed and F
calc are calculated structure-factor amplitudes.
The R free set used 5% of randomly chosen reflections.
