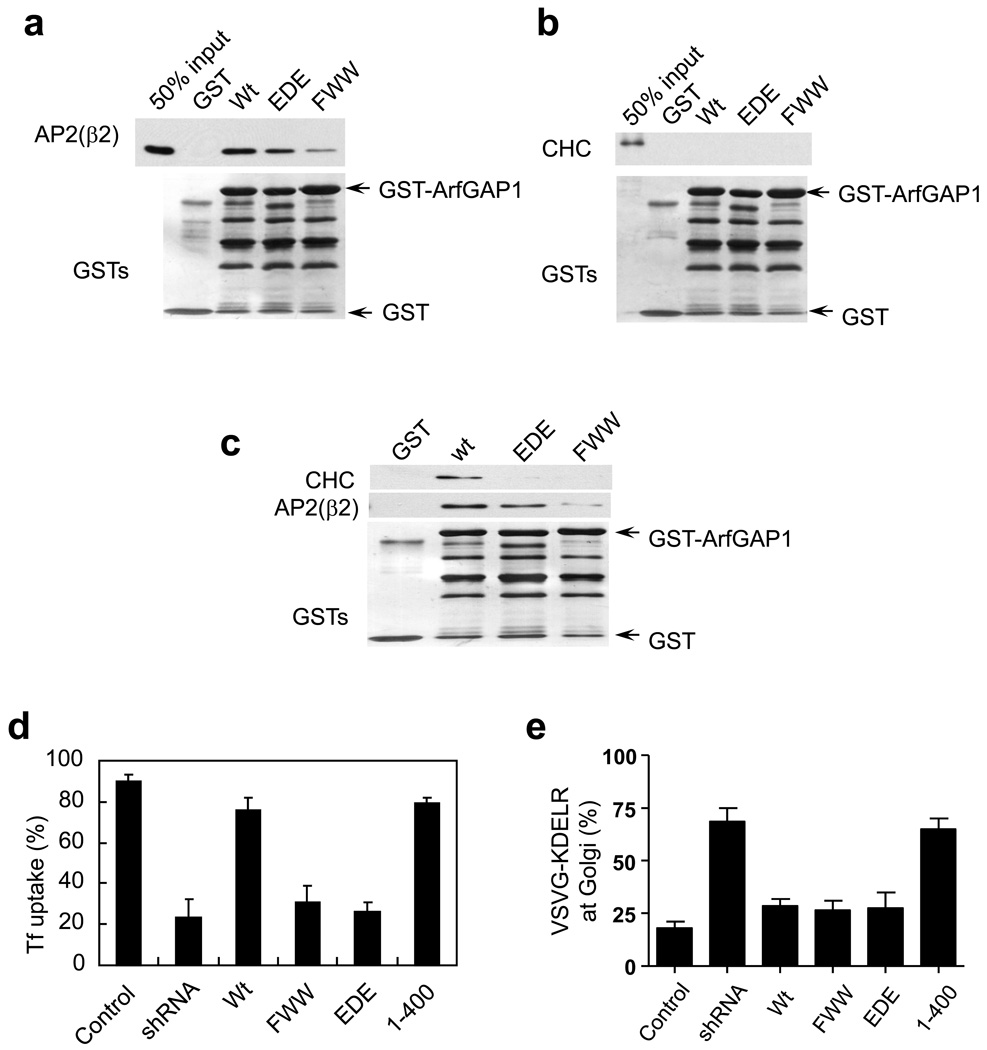
Figure 3. Disrupting interaction with either coatomer or AP-2 leads to selective disruption in transport pathways.
a. Pulldown assays to assess direct binding of ARFGAP1 to AP-2. The different forms of ARFGAP1 as GST fusion proteins were bound to beads, incubated with purified AP-2 adaptors, and then immunoblotted for proteins as indicated. GST fusion proteins were detected by Coomassie staining.
b. Pulldown assays to assess direct binding of ARFGAP1 to clathrin. The different forms of ARFGAP1 as GST fusion proteins were bound to beads, incubated with purified clathrin triskelia, and then immunoblotted for proteins as indicated. GST fusion proteins were detected by Coomassie staining.
c. Sequential incubations reveal that the AP-2 adaptor is needed to link ARFGAP1 to the clathrin triskelion. The different forms of ARFGAP1 as GST fusion proteins were bound to beads, and then incubated with purified AP-2 followed by incubation with purified clathrin. Beads were then analyzed by immunoblotted for proteins as indicated. GST fusion proteins were detected by Coomassie staining.
d. Rescue of defective Tf uptake induced by the depletion of ARFGAP1. BSC-1 cells that stably expressed shRNA against ARFGAP1 were transfected with different rat forms of ARFGAP1 as indicated. Uptake of biotin-Tf at 10 minutes was then quantified. The mean from three experiments with standard error is shown. Difference among conditions of shRNA, FWW and EDE are insignificant (p>0.05). Difference between this group and all other conditions are significant (p<0.05).
e. Rescue of defective COPI transport induced by the depletion of ARFGAP1. BSC-1 cells that stably expressed shRNA against ARFGAP1 were transfected with different rat forms of ARFGAP1 as indicated. The redistribution of VSVG-KDELR from the Golgi to the ER at 30 minutes was then quantified. The mean from three experiments with standard error is shown. Difference among conditions of Wt, FWW and EDE are insignificant (p>0.05). Difference between this group and conditions of shRNA and 1–400 are significant (p<0.05).