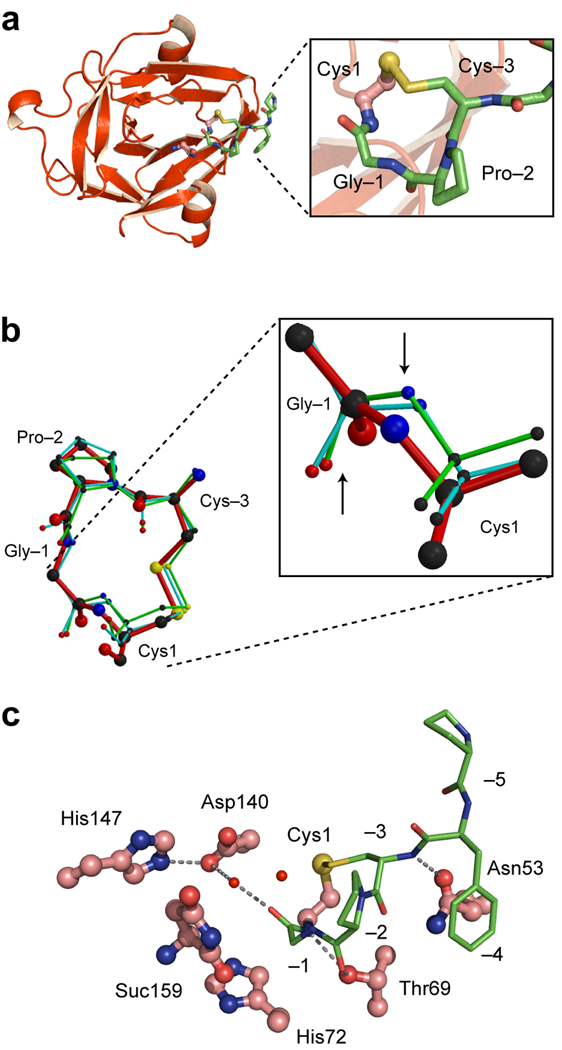
Figure 2.
Crystal structure of the CPGCDnaE intein. (a). Ribbon diagram showing the intein (red) and the N-terminal extein (green). Magnified view of the Cys1-to-Cys-3 disulfide loop is shown in the box (b). Conformation of the CPGC disulfide loop. Comparison of the structure of the disulfide loop in the CPGCDnaE intein (red) with the two other observations of this loop in the Protein Data Bank (1JBQ, green; 1M6Y, cyan). Magnified view shows Gly-Cys peptide bonds, with an upward arrow pointing to the carbonyl oxygen atoms and a downward arrow pointing toward the amide nitrogen atoms. (c). Active-site interactions in the CPGCDnaE intein (intein, red; extein, green). Thr69 of the conserved TXXH motif contacts the amide nitrogen of Cys1 and the carbonyl oxygen of Pro-2 while His72, also of the TXXH motif, does not interact directly with the N-extein. Asp140 makes a water-mediated contact with the −1 carbonyl and a hydrogen bond with His147, also a conserved residue. Suc 159 is the C-terminal succinimide.