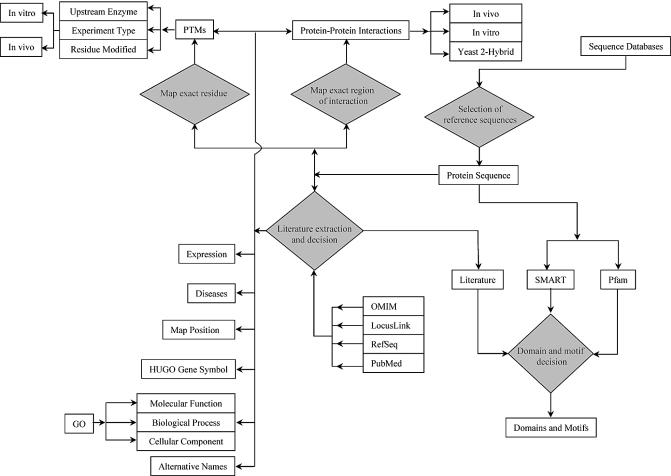
Figure 2.
A schematic outlining the basic steps in the annotation procedure. The annotations are carried out by trained biologists who critically read the published literature. The entries to be annotated are carefully selected from all the existing database entries by BLAST analysis (6) as well as manual inspection to provide the reference sequence. Interpretive annotation steps are represented by orange diamonds in the schematic. For instance, the annotator performs domain and motif analysis using the SMART (7) and Pfam (8) programs as well as by reading the literature. The OMIM database is used for disease annotations (9), and RefSeq and LocusLink (10) for sequences and links to other databases.