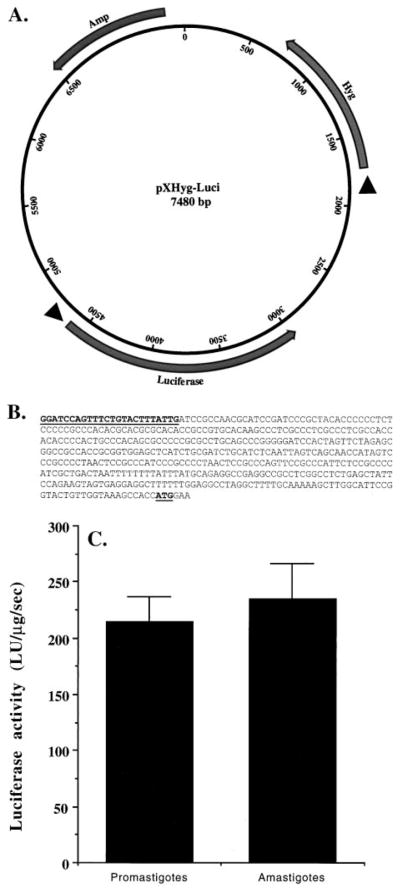
Fig. 3.
Stable expression of firefly luciferase in L. amazonensis promastigotes overexpressing T. brucei RNase H1. (A) Map of the plasmid pXHyg-Luci; Amp, Hyg, and Luciferase indicate the beta-lactamase, hygromycin phosphotransferase, and firefly luciferase genes. Arrowheads before the genes indicate Leishmania trans-splicing signals. These trans-splicing signals are needed for the expression of the genes cloned behind them. The orientations of the genes with respect to their promoter (Amp gene) or trans-splicing signal (Hyg or Luciferase genes) are indicated as solid arrows. (B) Nucleotide sequence (5′ to 3′) (453 bp) of the miniexon end of the 5′-RACE product from the mRNA of the recombinant cells using Mex2 and LUAS as primers. The Mex2 sequence and the luciferase mRNA translation start site (AUG) are boldfaced and underlined. (C) Expression of luciferase activity in the recombinant promastigotes and amastigotes. LU = light units. Results are means ± SEM (N = 6). The difference in luciferase activity between the amastigote and promastigote cell extracts was not statistically significant.