FIG 1 .
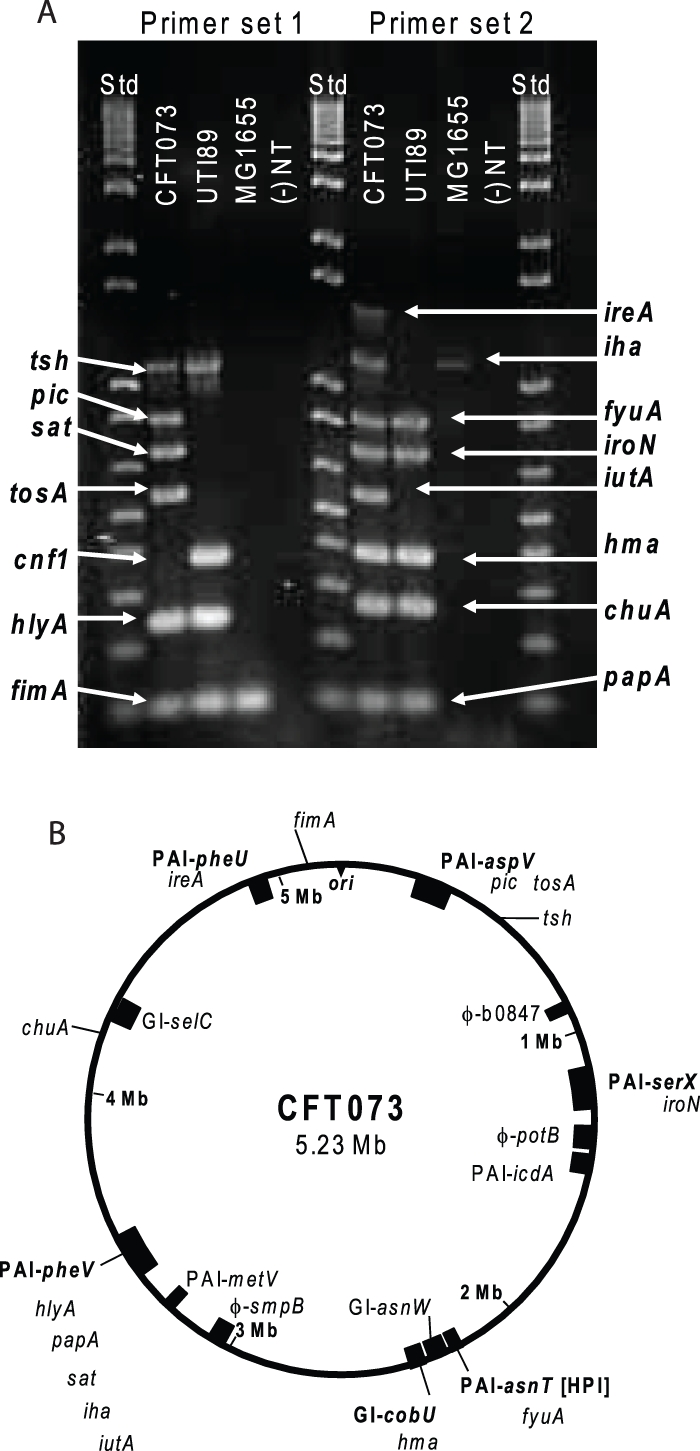
Virulence and fitness genes assessed in this study. (A) Multiplex PCR results using genomic DNA purified from CFT073, UTI89, and MG1655. Primers specific for each virulence or fitness gene were used for PCR amplification of gene fragments from a boiled lysate of the indicated E. coli strain. If there was no band observed at the expected size, the result was counted as negative for the presence of a gene (−). NT, no-template control samples; Std, DNA ladder standards. (B) Schematic of CFT073 genome with the location of each gene under study indicated. The origin of replication is indicated by ori. Previously identified pathogenicity-associated islands (PAI) or genomic islands (GI) containing one or more genes under study are indicated according to the nomenclature of Lloyd et al. (11). cnf1, which is not present in strain CFT073, is not shown. Phage insertion sites (φ). Three genes, tsh, chu, and fimA, are not found in a PAI or GI.
