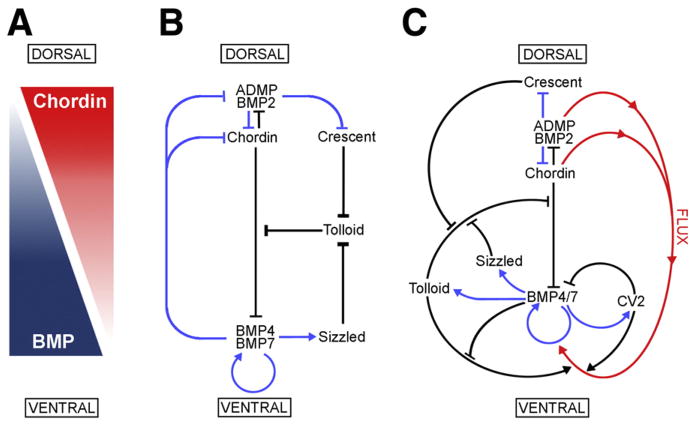
Fig. 7.
Model of the extracellular network of proteins that control D–V patterning. Crescent was shown here to be a component of the extracellular biochemical network that controls D–V patterning. Arrows in blue represent transcriptional regulation by BMPs, arrows in black symbolize direct protein–protein interactions, and the red arrows indicate flux of Chordin/ADMP/BMP from the dorsal toward the ventral side of the embryo. Each protein–protein interaction indicated here is supported by biochemical and embryological studies in Xenopus. (A) The BMP/Chordin gradient plays a central role in D–V patterning. (B) Tolloid metalloproteinases are inhibited by Crescent from the dorsal and Sizzled from the ventral side. (C) The extracellular biochemical pathway of D–V development in Xenopus embryos, including the flux of Chordin/BMP complexes from dorsal to ventral. Two important regulators of the Chordin/BMP pathway have been omitted for simplicity in this diagram. One is Ont-1, an adaptor Olfactomedin-related protein that binds both Chordin and Tolloid, which facilitates Chordin proteolysis (Inomata et al., 2008). The other regulator is Twisted-gastrulation (Tsg), a protein that binds both BMP and Chordin, facilitating BMP signaling, the binding of BMP to Chordin, and the cleavage of Chordin by Tolloids (De Robertis and Kuroda, 2004; Little and Mullins, 2006).