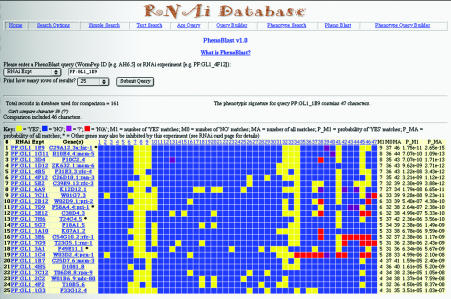
Figure 2.
An example of a ‘PhenoBlast’ result page. The phenotypic signature of the query is shown in the first row of the result list, followed by the 24 best matches in the database. In this example, the query (RNAi experiment ‘PF:GL1_1E9’, corresponding to gene ‘lig-1’) is thought to be involved in DNA synthesis, as are many top-ranking genes (based on sequence similarity to genes of known function in other organisms). Wild-type states are represented by a blue square while mutant phenotypes are represented by yellow squares. Red and purple squares represent cases where the phenotype state for that character is, respectively, missing or unknowable under these assay conditions. Results are ranked by similarity to the query signature and are sorted lexicographically based on the number of shared phenotypic defects (M1), the total number of shared phenotypic characters (MA) and the probability of observing this combination of shared defects (P_M1) or all shared characters (P_MA) in the population of experiments in the database. Results with the same number of shared characters are thus ranked in a weighted fashion that is inversely proportional to the frequency of character states in the population. Each result row is linked to the corresponding RNAi Experiment and Gene Product card pages.