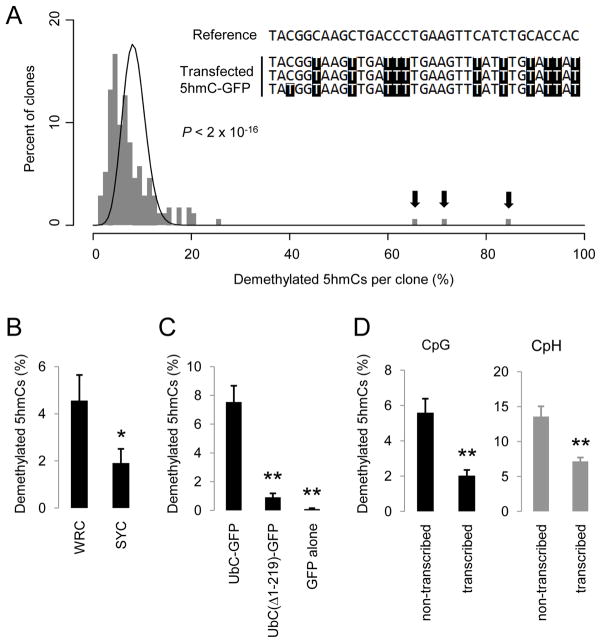
Figure 5. 5hmC Demethylation Recapitulates Known Properties of AID-Catalysed Deamination.
(A) A histogram of clone distribution of the percentage of demethylated 5hmCs. A Poisson distribution with λ = 8.4% is shown as a black line. Arrows indicate 3 highly processively demethylated 5hmC-GFP clones. A small portion of the reference sequence and the bisulfite sequencing results for these 3 clones are shown in the inset.
(B) Occurrence of single demethylated 5hmCs at WRC and SYC motifs. WRC motif: W = A/T; R = A/G; SYC motif: S = C/G; Y =C/T. Only demethylated 5hmCs that are at least 5 nucleotides away from any other demethylated 5hmCs are analyzed. Values represent mean ± SEM (n = 3; *: P < 0.05; Student t-test).
(C) Demethylation of promoter-truncated 5hmC-GFP variants. The same region in the GFP open-reading frame is bisulfite sequenced for all variants. Values represent mean ± SEM (n = 3; **: P < 0.01; Student t-test).
(D) Demethylation of 5hmCs in the CpG (left) and CpH (right) contexts on two opposite strands. Demethylated 5hmCs of the forward (untranscribed) strand and the reverse (transcribed) strand are represented by C-to-T and G-to-A transitions in bisulfite sequencing (Figure S5C), respectively. Values represent mean ± SEM (n = 3; **: P < 0.01; Student’s t-test).