Figure 3.
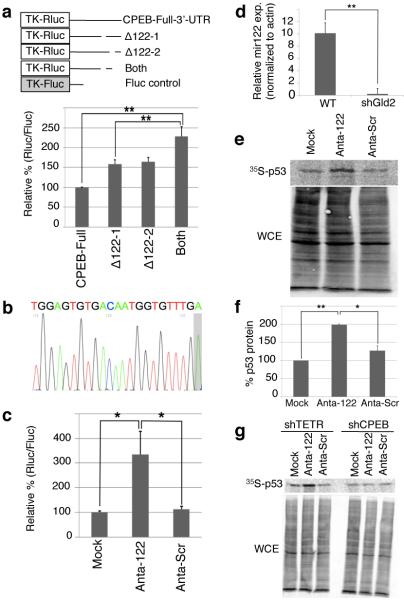
miR-122 activates p53 mRNA translation by repressing CPEB. (a) Gld2 depleted fibroblasts were transduced with firefly and Renilla luciferase with CPEB full length or deletion mutant 3’UTRs lacking putative miR-122 sites (Supplementary Fig. 2). The data are expressed as in Fig 2; in all panels, n = 3 and the bars refer to s.e.m. and one or two asterisks refer to statistical significance (Students t test) at the p<0.05 or p<0.01 levels, respectively. (b) Sequence of miR-122 from fibroblasts; a non-templated adenosine is shaded. (c) Fibroblasts expressing firefly and Renilla luciferase containing the CPEB 3’ UTR were electroporated with miR-122 (Anta-122), scrambled, (Anta-Scr), or no LNA antagomir (Mock); data are expressed as in Figure 2. (d) Quantitative RTPCR for miR-122 in cells expressing GFP (WT) or shGld2. (e and f) Immunoprecipitation of 35S-methonine-labeled p53 from MG132-treated cells transduced with no (Mock-GFP), miR-122 (Anta-122), or scrambled (Anta-Scram) LNA antagomirs. WCE refers to whole cell lysate. (g) Fibroblasts were treated as in panels e-g after first expressing either TET repressor (shTETR, a control) or CPEB shRNA.