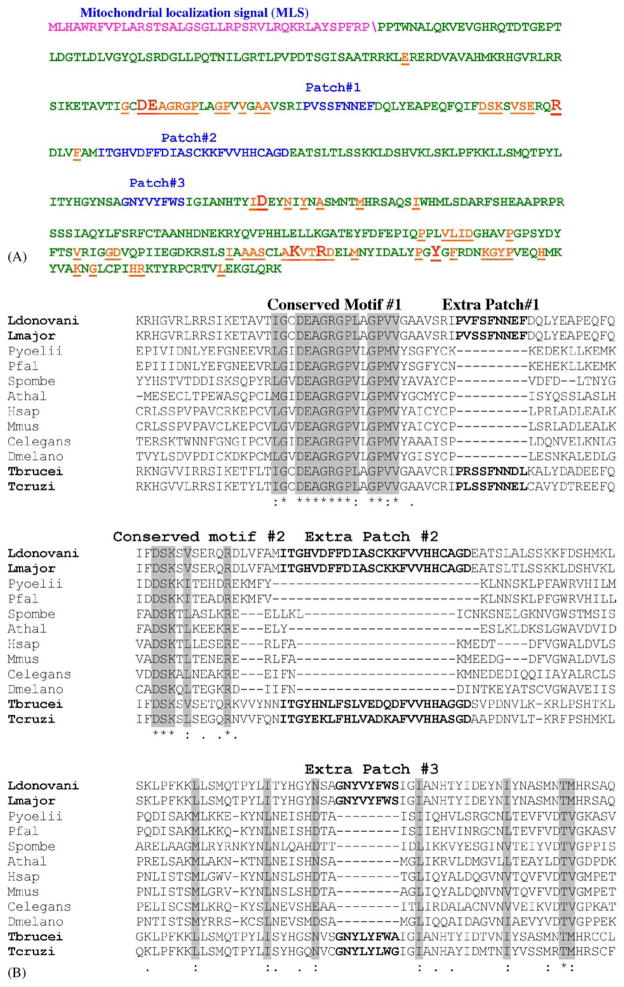
Fig. 1.
Analysis of LRNase HIIC amino acid sequence. (A) The amino acid sequence of LRNase HIIC highlighting the MLS (purple)\shows the predicted cleavage site, conserved amino acids (orange) and the extra patches of sequences (blue). The conserved amino acid residues that are essential for the enzymatic activity of the protein are shown in larger fonts. (B) ClustalW alignment of part of amino acid sequences of L. donovani LRNase HIIC (residues 119–298) with corresponding homologous RNase HII sequences from selected eukaryotes to show the relative location of the extra patches of amino acid sequences. The names of the organisms and the accession numbers of their RNase HII proteins are as follows: Ldonovani: L. donovani, AAL57850; Lmajor: L. major, AAL32059; Pyoellii: Plasmodium yoellii, EAA18794; Pfal: Plasmodium falciparum, NP 703896; T. brucei, AAD53318; Spombe: Schizosaccharomyces pombe, Q10236; Athal: Arabidopsis thalina, AAO64017; Hsap: Homo sapiens, AAQ64005; Mmus: Mus musculus, NP 081463; Celegans: Caenorhabditis elegans, CAC70103; and Dmelano: Drosophila melanogaster, Q9VPP5. The sequences for T. brucei and T. cruzi RNase HIIC were retrieved from the respective GeneDB (see the footnote of Table 2).