Fig. 5.
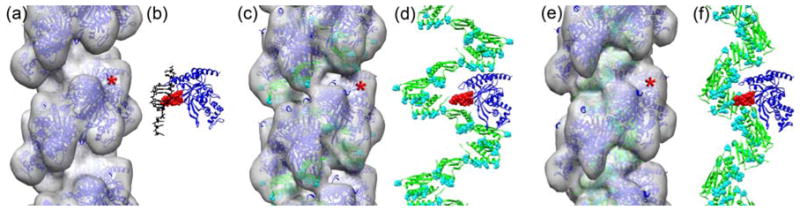
In the atomic model of the pure RecA filament (a, blue ribbons) the dsDNA molecule (b, black ribbons) is proximal to the L2-loop of the RecA (b, red spheres). When DinI binds to RecA in the CTD mode (c,d, green ribbons) the six Asp/Glu residues in the C-terminal helix that form an extended negatively charged ridge36 (c,d, cyan spheres) are distal from the L2-loop of RecA (d, red spheres). In the atomic model of the groove mode the negatively charged C-terminal helix forms a continuous helical pattern (e,f,, cyan spheres) flanking the L2 region of RecA (f, red spheres). The position of the single RecA protomer (blue ribbons in b,d,f) is marked with red asterisks in the filament models (a,c,e).
