Abstract
The title compound, C17H12N2O3, was synthesized by the reaction of 2-hydroxy-1-naphthaldehyde with 4-nitrobenzenamine. These condense to form the Schiff base, which crystallizes in the zwitterionic form. In the structure, the keto–amino tautomer has a fairly short intramolecular N—H⋯O hydrogen bond between the 2-naphthalenone and amino groups, with electron delocalization. The molecule is essentially planar, with a dihedral angle of 1.96 (3)° between the ring systems. In the crystal, the molecules are linked via intermolecular C—H⋯O hydrogen bonds, forming a layer parallel to (101).
Related literature
For background to Schiff base compounds, see: Fan et al. (2007 ▶); Kim et al. (2005 ▶); Nimitsiriwat et al. (2004 ▶). For the pharmaceutical and medicinal activity of Schiff bases, see: Chen et al. (1997 ▶); Dao et al. (2000 ▶); Ren et al. (2002 ▶); Sriram et al. (2006 ▶); Karthikeyan et al. (2006 ▶). For Schiff bases in coordination chemistry, see: Ali et al. (2008 ▶); Kargar et al. (2009 ▶); Yeap et al. (2009 ▶). For related structures, see: Fun et al. (2009 ▶); Nadeem et al. (2009 ▶); Eltayeb et al. (2008 ▶). For standard bond lengths see: Allen, (2002 ▶). 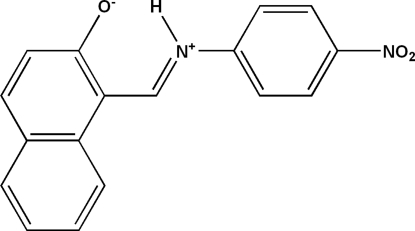
Experimental
Crystal data
C17H12N2O3
M r = 292.29
Monoclinic,

a = 8.0503 (6) Å
b = 12.8174 (9) Å
c = 13.1833 (10) Å
β = 97.271 (5)°
V = 1349.37 (17) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 296 K
0.15 × 0.06 × 0.04 mm
Data collection
Bruker SMART CCD area-detector diffractometer
44074 measured reflections
7946 independent reflections
3658 reflections with I > 2σ(I)
R int = 0.074
Refinement
R[F 2 > 2σ(F 2)] = 0.059
wR(F 2) = 0.190
S = 0.96
7946 reflections
207 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.53 e Å−3
Δρmin = −0.26 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SIR2002 (Burla et al., 2003 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and DIAMOND (Brandenburg & Berndt, 2001 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811012359/bq2290sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811012359/bq2290Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2N⋯O3 | 1.09 (2) | 1.57 (2) | 2.5287 (15) | 143 (2) |
| C5—H5⋯O2i | 0.93 | 2.59 | 3.5136 (16) | 173 |
| C16—H16⋯O2i | 0.93 | 2.53 | 3.4455 (17) | 169 |
Symmetry code: (i)  .
.
Acknowledgments
This work was supported by the Unité de Recherche de Chimie de l’Environnement et Moléculaire Structurale, CHEMS, Université Mentouri-Constantine, Algeria and the Laboratoire de Chimie de Coordination, Toulouse, France. Thanks are due to the MESRS (Ministére de l’Enseignement Supérieur et de la Recherche Scientifique - Algérie) for financial support.
supplementary crystallographic information
Comment
Schiff base compounds have been widely investigated over a century (Fan et al., 2007; Kim et al., 2005; Nimitsiriwat et al., 2004). Some of the compounds have been found to have pharmaceutical and medicinal fields (Chen et al., 1997; Ren et al., 2002; Dao et al., 2000; Sriram et al., 2006; Karthikeyan et al., 2006). They are also used as versatile ligands in coordination chemistry (Ali et al., 2008; Kargar et al., 2009; Yeap et al., 2009).
As part of our ongoing studies of Schiff base complexes and derivatives we report here synthesis and the crystal structure of the title compound, obtained by the reaction of 2-hydroxy-1-naphthaldehyde with 4-nitroaniline, which crystallized in a zwitterionic form with cationic iminium and anionic naphtholate group.
The molecular structure of (I), and the atomic numbering used, is illustrated in Fig. 1. All bond distances and angles are within the ranges of accepted values (CSD, Allen, 2002) and in literature (Fun et al., 2009; Nadeem et al., 2009; Eltayeb et al., 2008).
The main molecule is essentially planar with an rms deviation of 0.0350 Å, and the crystal structure exhibit alternating layers parallel to (101) plane (Fig. 2). In the crystal, molecules are linked via intermolecular C—H···O hydrogen bonds to form a two-dimensional layers parallel to (101) (Table 1, Fig. 3) and additional stabilization within these layers is provided by N—O···π and π···π stacking interactions. These interaction bonds link the molecules within the layers and also link the layers together and reinforcing the cohesion of the structure. An intramolecular N—H···O hydrogen bond occurs.
Experimental
The title compound, (I), was prepared by refluxing a mixture of a solution containing (0.1 mmol) of 2-hydroxy-1-naphthaldehyde and (0.1 mmol) of 4-nitrobenzenamine in 20 ml methanol. The reaction mixture was stirred for 1 h under reflux. Microcrystals of (I) were obtained by allowing the clear solution to stand overnight. The powder product was dissolved and recrystallized from DMSO solution. Some red crystals were carefully isolated under polarizing microscope for analysis by x-ray diffraction.
Refinement
H7 and H2N were located in difference Fourier maps and refined isotropically. The remaining H atoms were localized on Fourier maps but introduced in calculated positions and treated as riding on their parent atoms (Caryl) with Caryl—Haryl=0.95Å and Uiso(Haryl)=1.2Ueq(Caryl).
Figures
Fig. 1.
(Farrugia, 1997) The asymmetric unit of the title compound with the atomic labeling scheme. Displacement are drawn at the 50% probability level. Hydrogen bond shown as dashed line.
Fig. 2.
(Brandenburg, 2001) A diagram of the layered crystal packing in (I), viewed down the b axis, showing layers parallel to (101).
Fig. 3.
(Brandenburg, 2001) A part of crystal packing of (I) showing hydrogen bond connections in the same layer as dashed line.
Crystal data
| C17H12N2O3 | F(000) = 608 |
| Mr = 292.29 | Dx = 1.439 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 5160 reflections |
| a = 8.0503 (6) Å | θ = 3.0–30.1° |
| b = 12.8174 (9) Å | µ = 0.10 mm−1 |
| c = 13.1833 (10) Å | T = 296 K |
| β = 97.271 (5)° | Needle, red |
| V = 1349.37 (17) Å3 | 0.15 × 0.06 × 0.04 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3658 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.074 |
| graphite | θmax = 39.3°, θmin = 3.0° |
| φ and ω scans | h = −14→12 |
| 44074 measured reflections | k = −20→22 |
| 7946 independent reflections | l = −20→23 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.190 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.96 | w = 1/[σ2(Fo2) + (0.0963P)2] where P = (Fo2 + 2Fc2)/3 |
| 7946 reflections | (Δ/σ)max = 0.001 |
| 207 parameters | Δρmax = 0.53 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O3 | 0.88419 (14) | −0.08384 (7) | 0.31991 (8) | 0.0392 (3) | |
| O2 | 0.44266 (15) | −0.40551 (8) | 0.79254 (8) | 0.0426 (3) | |
| O1 | 0.49601 (17) | −0.51766 (7) | 0.67875 (9) | 0.0500 (3) | |
| N2 | 0.74356 (13) | −0.11314 (7) | 0.47852 (8) | 0.0264 (2) | |
| N1 | 0.49499 (15) | −0.42784 (8) | 0.71179 (9) | 0.0320 (2) | |
| C17 | 0.82974 (14) | 0.16828 (9) | 0.44636 (9) | 0.0234 (2) | |
| C8 | 0.82177 (14) | 0.05641 (9) | 0.42782 (9) | 0.0234 (2) | |
| C6 | 0.55083 (16) | −0.24287 (9) | 0.68580 (10) | 0.0274 (2) | |
| H6 | 0.5057 | −0.2275 | 0.7456 | 0.033* | |
| C1 | 0.55862 (16) | −0.34487 (9) | 0.65258 (10) | 0.0259 (2) | |
| C7 | 0.75161 (14) | −0.01080 (9) | 0.49403 (10) | 0.0245 (2) | |
| C2 | 0.62367 (17) | −0.37057 (9) | 0.56335 (10) | 0.0300 (3) | |
| H2 | 0.6267 | −0.4396 | 0.5421 | 0.036* | |
| C12 | 0.90181 (15) | 0.23306 (9) | 0.37664 (10) | 0.0273 (2) | |
| C16 | 0.76748 (16) | 0.21715 (9) | 0.52907 (10) | 0.0293 (3) | |
| H16 | 0.721 | 0.1766 | 0.5768 | 0.035* | |
| C3 | 0.68369 (16) | −0.29201 (9) | 0.50680 (10) | 0.0290 (3) | |
| H3 | 0.7278 | −0.3079 | 0.4468 | 0.035* | |
| C5 | 0.61127 (16) | −0.16388 (9) | 0.62877 (10) | 0.0265 (2) | |
| H5 | 0.6069 | −0.0949 | 0.6501 | 0.032* | |
| C13 | 0.90891 (17) | 0.34145 (10) | 0.39091 (12) | 0.0355 (3) | |
| H13 | 0.9569 | 0.383 | 0.3446 | 0.043* | |
| C10 | 0.96505 (18) | 0.08390 (11) | 0.27475 (11) | 0.0352 (3) | |
| H10 | 1.0122 | 0.0574 | 0.2193 | 0.042* | |
| C4 | 0.67859 (14) | −0.18811 (9) | 0.53937 (9) | 0.0239 (2) | |
| C9 | 0.88945 (16) | 0.01310 (9) | 0.34053 (10) | 0.0280 (2) | |
| C14 | 0.84615 (17) | 0.38702 (10) | 0.47216 (13) | 0.0379 (3) | |
| H14 | 0.8517 | 0.459 | 0.4812 | 0.045* | |
| C15 | 0.77392 (17) | 0.32417 (10) | 0.54105 (12) | 0.0346 (3) | |
| H15 | 0.7296 | 0.3547 | 0.5957 | 0.041* | |
| C11 | 0.96888 (17) | 0.18704 (10) | 0.29171 (11) | 0.0336 (3) | |
| H11 | 1.0168 | 0.2302 | 0.2467 | 0.04* | |
| H7 | 0.704 (2) | 0.0100 (13) | 0.5544 (14) | 0.043 (5)* | |
| H2N | 0.798 (3) | −0.1327 (18) | 0.4091 (18) | 0.080 (7)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0597 (6) | 0.0256 (4) | 0.0362 (5) | −0.0031 (4) | 0.0217 (5) | −0.0042 (4) |
| O2 | 0.0637 (7) | 0.0342 (5) | 0.0348 (5) | −0.0030 (5) | 0.0249 (5) | 0.0036 (4) |
| O1 | 0.0847 (9) | 0.0207 (4) | 0.0502 (7) | −0.0069 (5) | 0.0308 (6) | 0.0002 (4) |
| N2 | 0.0325 (5) | 0.0198 (4) | 0.0283 (5) | −0.0011 (4) | 0.0097 (4) | 0.0016 (4) |
| N1 | 0.0419 (6) | 0.0241 (5) | 0.0322 (6) | −0.0016 (4) | 0.0134 (5) | 0.0031 (4) |
| C17 | 0.0224 (5) | 0.0216 (5) | 0.0260 (6) | −0.0003 (4) | 0.0023 (4) | 0.0025 (4) |
| C8 | 0.0253 (5) | 0.0215 (5) | 0.0235 (5) | −0.0003 (4) | 0.0044 (4) | 0.0014 (4) |
| C6 | 0.0332 (6) | 0.0226 (5) | 0.0282 (6) | −0.0006 (4) | 0.0111 (5) | −0.0012 (4) |
| C1 | 0.0320 (6) | 0.0206 (4) | 0.0264 (6) | −0.0007 (4) | 0.0089 (5) | 0.0015 (4) |
| C7 | 0.0256 (5) | 0.0212 (5) | 0.0271 (6) | −0.0002 (4) | 0.0052 (4) | 0.0017 (4) |
| C2 | 0.0395 (7) | 0.0196 (5) | 0.0335 (6) | 0.0000 (4) | 0.0148 (5) | −0.0022 (4) |
| C12 | 0.0257 (5) | 0.0236 (5) | 0.0329 (6) | −0.0022 (4) | 0.0054 (5) | 0.0037 (4) |
| C16 | 0.0334 (6) | 0.0251 (5) | 0.0302 (6) | −0.0001 (5) | 0.0073 (5) | −0.0012 (4) |
| C3 | 0.0369 (6) | 0.0222 (5) | 0.0306 (6) | −0.0001 (4) | 0.0151 (5) | −0.0018 (4) |
| C5 | 0.0326 (6) | 0.0193 (4) | 0.0290 (6) | −0.0019 (4) | 0.0097 (5) | −0.0026 (4) |
| C13 | 0.0334 (6) | 0.0245 (5) | 0.0499 (9) | −0.0044 (5) | 0.0108 (6) | 0.0053 (5) |
| C10 | 0.0471 (8) | 0.0325 (6) | 0.0291 (6) | −0.0030 (5) | 0.0164 (6) | 0.0009 (5) |
| C4 | 0.0268 (5) | 0.0201 (4) | 0.0259 (6) | −0.0012 (4) | 0.0075 (4) | 0.0005 (4) |
| C9 | 0.0334 (6) | 0.0257 (5) | 0.0259 (6) | −0.0012 (4) | 0.0076 (5) | −0.0006 (4) |
| C14 | 0.0357 (7) | 0.0220 (5) | 0.0565 (9) | −0.0029 (5) | 0.0079 (7) | −0.0021 (6) |
| C15 | 0.0367 (7) | 0.0260 (6) | 0.0413 (8) | 0.0011 (5) | 0.0059 (6) | −0.0060 (5) |
| C11 | 0.0382 (7) | 0.0319 (6) | 0.0329 (7) | −0.0054 (5) | 0.0129 (6) | 0.0046 (5) |
Geometric parameters (Å, °)
| O3—C9 | 1.2714 (15) | C2—H2 | 0.93 |
| O2—N1 | 1.2274 (14) | C12—C13 | 1.4021 (17) |
| O1—N1 | 1.2312 (14) | C12—C11 | 1.4304 (18) |
| N2—C7 | 1.3279 (15) | C16—C15 | 1.3810 (17) |
| N2—C4 | 1.3951 (14) | C16—H16 | 0.93 |
| N2—H2N | 1.09 (2) | C3—C4 | 1.4015 (16) |
| N1—C1 | 1.4504 (14) | C3—H3 | 0.93 |
| C17—C16 | 1.4039 (16) | C5—C4 | 1.3931 (16) |
| C17—C12 | 1.4163 (15) | C5—H5 | 0.93 |
| C17—C8 | 1.4546 (16) | C13—C14 | 1.372 (2) |
| C8—C7 | 1.3957 (15) | C13—H13 | 0.93 |
| C8—C9 | 1.4450 (16) | C10—C11 | 1.3405 (18) |
| C6—C1 | 1.3827 (17) | C10—C9 | 1.4421 (17) |
| C6—C5 | 1.3855 (16) | C10—H10 | 0.93 |
| C6—H6 | 0.93 | C14—C15 | 1.395 (2) |
| C1—C2 | 1.3866 (16) | C14—H14 | 0.93 |
| C7—H7 | 0.962 (19) | C15—H15 | 0.93 |
| C2—C3 | 1.3763 (16) | C11—H11 | 0.93 |
| C7—N2—C4 | 127.33 (10) | C17—C16—H16 | 119.3 |
| C7—N2—H2N | 109.9 (12) | C2—C3—C4 | 120.22 (11) |
| C4—N2—H2N | 122.8 (12) | C2—C3—H3 | 119.9 |
| O2—N1—O1 | 122.91 (11) | C4—C3—H3 | 119.9 |
| O2—N1—C1 | 118.62 (10) | C6—C5—C4 | 119.81 (10) |
| O1—N1—C1 | 118.47 (10) | C6—C5—H5 | 120.1 |
| C16—C17—C12 | 117.28 (11) | C4—C5—H5 | 120.1 |
| C16—C17—C8 | 123.91 (10) | C14—C13—C12 | 120.93 (12) |
| C12—C17—C8 | 118.81 (10) | C14—C13—H13 | 119.5 |
| C7—C8—C9 | 118.96 (10) | C12—C13—H13 | 119.5 |
| C7—C8—C17 | 121.07 (10) | C11—C10—C9 | 121.53 (12) |
| C9—C8—C17 | 119.96 (10) | C11—C10—H10 | 119.2 |
| C1—C6—C5 | 119.07 (10) | C9—C10—H10 | 119.2 |
| C1—C6—H6 | 120.5 | C5—C4—N2 | 123.17 (10) |
| C5—C6—H6 | 120.5 | C5—C4—C3 | 120.04 (10) |
| C6—C1—C2 | 122.04 (11) | N2—C4—C3 | 116.79 (10) |
| C6—C1—N1 | 119.32 (10) | O3—C9—C10 | 119.44 (11) |
| C2—C1—N1 | 118.64 (10) | O3—C9—C8 | 122.68 (11) |
| N2—C7—C8 | 121.95 (11) | C10—C9—C8 | 117.87 (11) |
| N2—C7—H7 | 112.6 (10) | C13—C14—C15 | 119.19 (12) |
| C8—C7—H7 | 125.4 (10) | C13—C14—H14 | 120.4 |
| C3—C2—C1 | 118.81 (11) | C15—C14—H14 | 120.4 |
| C3—C2—H2 | 120.6 | C16—C15—C14 | 120.79 (12) |
| C1—C2—H2 | 120.6 | C16—C15—H15 | 119.6 |
| C13—C12—C17 | 120.46 (11) | C14—C15—H15 | 119.6 |
| C13—C12—C11 | 120.03 (11) | C10—C11—C12 | 122.29 (11) |
| C17—C12—C11 | 119.50 (11) | C10—C11—H11 | 118.9 |
| C15—C16—C17 | 121.34 (11) | C12—C11—H11 | 118.9 |
| C15—C16—H16 | 119.3 | ||
| C16—C17—C8—C7 | −0.89 (19) | C1—C6—C5—C4 | 0.0 (2) |
| C12—C17—C8—C7 | −179.83 (12) | C17—C12—C13—C14 | −0.2 (2) |
| C16—C17—C8—C9 | −179.95 (12) | C11—C12—C13—C14 | −179.64 (14) |
| C12—C17—C8—C9 | 1.11 (18) | C6—C5—C4—N2 | −179.26 (12) |
| C5—C6—C1—C2 | −0.6 (2) | C6—C5—C4—C3 | 0.6 (2) |
| C5—C6—C1—N1 | −179.78 (12) | C7—N2—C4—C5 | −0.3 (2) |
| O2—N1—C1—C6 | −3.2 (2) | C7—N2—C4—C3 | 179.85 (12) |
| O1—N1—C1—C6 | 176.80 (13) | C2—C3—C4—C5 | −0.6 (2) |
| O2—N1—C1—C2 | 177.56 (13) | C2—C3—C4—N2 | 179.28 (12) |
| O1—N1—C1—C2 | −2.4 (2) | C11—C10—C9—O3 | 177.94 (14) |
| C4—N2—C7—C8 | 179.22 (12) | C11—C10—C9—C8 | −1.8 (2) |
| C9—C8—C7—N2 | −0.77 (19) | C7—C8—C9—O3 | 1.8 (2) |
| C17—C8—C7—N2 | −179.84 (11) | C17—C8—C9—O3 | −179.16 (12) |
| C6—C1—C2—C3 | 0.6 (2) | C7—C8—C9—C10 | −178.46 (12) |
| N1—C1—C2—C3 | 179.80 (12) | C17—C8—C9—C10 | 0.62 (19) |
| C16—C17—C12—C13 | −0.20 (19) | C12—C13—C14—C15 | −0.2 (2) |
| C8—C17—C12—C13 | 178.81 (12) | C17—C16—C15—C14 | −1.4 (2) |
| C16—C17—C12—C11 | 179.27 (12) | C13—C14—C15—C16 | 1.0 (2) |
| C8—C17—C12—C11 | −1.71 (18) | C9—C10—C11—C12 | 1.3 (2) |
| C12—C17—C16—C15 | 0.98 (19) | C13—C12—C11—C10 | −179.98 (14) |
| C8—C17—C16—C15 | −177.98 (13) | C17—C12—C11—C10 | 0.5 (2) |
| C1—C2—C3—C4 | 0.0 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2N···O3 | 1.09 (2) | 1.57 (2) | 2.5287 (15) | 143 (2) |
| C5—H5···O2i | 0.93 | 2.59 | 3.5136 (16) | 173 |
| C16—H16···O2i | 0.93 | 2.53 | 3.4455 (17) | 169 |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BQ2290).
References
- Ali, H. M., Mohamed Mustafa, M. I., Rizal, M. R. & Ng, S. W. (2008). Acta Cryst. E64, m718–m719. [DOI] [PMC free article] [PubMed]
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Brandenburg, K. & Berndt, M. (2001). DIAMOND Crystal Impact, Bonn, Germany.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Chen, H. Q., Hall, S., Zheng, B. & Rhodes, J. (1997). Biodrugs, 7, 217–231. [DOI] [PubMed]
- Dao, V.-T., Gaspard, C., Mayer, M., Werner, G. H., Nguyen, S. N. & Michelot, R. J. (2000). Eur. J. Med. Chem. 35, 805–813. [DOI] [PubMed]
- Eltayeb, N. E., Teoh, S. G., Chantrapromma, S., Fun, H.-K. & Adnan, R. (2008). Acta Cryst. E64, o576–o577. [DOI] [PMC free article] [PubMed]
- Fan, Y. H., He, X. T., Bi, C. F., Guo, F., Bao, Y. & Chen, R. (2007). Russ. J. Coord. Chem. 33, 535–538.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Fun, H.-K., Kia, R., Vijesh, A. M. & Isloor, A. M. (2009). Acta Cryst. E65, o349–o350. [DOI] [PMC free article] [PubMed]
- Kargar, H., Jamshidvand, A., Fun, H.-K. & Kia, R. (2009). Acta Cryst. E65, m403–m404. [DOI] [PMC free article] [PubMed]
- Karthikeyan, M. S., Prasad, D. J., Poojary, B., Bhat, K. S., Holla, B. S. & Kumari, N. S. (2006). Bioorg. Med. Chem. 14, 7482–7489. [DOI] [PubMed]
- Kim, H.-J., Kim, W., Lough, A. J., Kim, B. M. & Chin, J. (2005). J. Am. Chem. Soc. 127, 16776–16777. [DOI] [PubMed]
- Nadeem, S., Shah, M. R. & VanDerveer, D. (2009). Acta Cryst. E65, o897. [DOI] [PMC free article] [PubMed]
- Nimitsiriwat, N., Marshall, E. L., Gibson, V. C., Elsegood, M. R. J. & Dale, S. H. (2004). J. Am. Chem. Soc. 126, 13598–13599. [DOI] [PubMed]
- Ren, S., Wang, R., Komatsu, K., Bonaz-Krause, P., Zyrianov, Y., McKenna, C. E., Csipke, C., Tokes, Z. A. & Lien, E. J. (2002). J. Med. Chem. 45, 410–419. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sriram, D., Yogeeswari, P., Myneedu, N. S. & Saraswat, V. (2006). Bioorg. Med. Chem. Lett. 16, 2127–2129. [DOI] [PubMed]
- Yeap, C. S., Kia, R., Kargar, H. & Fun, H.-K. (2009). Acta Cryst. E65, m570–m571. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811012359/bq2290sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811012359/bq2290Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





