Abstract
Anterior head segmentation is governed by different regulatory mechanisms than those that control trunk segmentation in Drosophila. For segment polarity genes, both initial mode of activation as well as cross-regulatory interactions among them differ from the typical genetic circuitry in the trunk and are unique for each of the procephalic segments. In order to better understand the segment-specific gene network responsible for the procephalic expression of the earliest active segment polarity genes wingless and hedgehog, we started to identify and analyze cis-regulatory DNA elements of these genes. For hedgehog, we could identify a cis-regulatory element, ic-CRE, that mediates expression specifically in the posterior part of the intercalary segment and requires promoter-specific interaction for its function. The intercalary stripe is the last part of the metameric hedgehog expression pattern that appears during embryonic development, which probably reflects the late and distinct establishment of this segment. The identification of a cis-regulatory element that is specific for one head segment supports the mutant-based observation that the expression of segment polarity genes is governed by a unique gene network in each of the procephalic segments. This provides further indication that the anterior-most head segments represent primary segments, which are set up independently, in contrast to the secondary segments of the trunk, which resemble true repetitive units.
Keywords: Enhancer dissection, Enhancer–promoter interaction, Head development, Metamerization, Segmentation
Introduction
Comparisons among distantly related insects suggest that head and trunk segmentation may have followed different paths during arthropod evolution (Minelli 2001; Tautz 2004). The mechanism of trunk segmentation found in the vinegar fly Drosophila melanogaster represents a relatively recent adjustment to its rapid embryonic development, whereas the mode of head development has been proposed to correspond to a phylogenetically more ancient mechanism. Comparisons with vertebrates even suggest that the origin of cephalization may be common for all Bilateria (Reichert and Simeone 1999). However, due to complex morphogenetic movements during embryonic head development of the acephalic Drosophila maggot, understanding head segmentation has not progressed as far as trunk segmentation in this species.
The insect head is built by several segment primordia that fuse to form the rigid head capsule with limbs that are adapted to different roles in orientation and feeding. The anterior head — the procephalon — consists of the ocular (protocerebral) region whose segmental status is debated, the antennal (deuterocerebral) and the intercalary (tritocerebral) segment (Scholtz and Edgecombe 2006). The gnathocephalon — mandibular, maxillary, and labial segments — constitutes the posterior portion of the insect head. In Drosophila, the labial and maxillary gnathal segments are patterned like the trunk segments according to the well-established hierarchical segmentation cascade involving maternal coordinate genes, gap, pair-rule, and segment polarity genes (St Johnston and Nüsslein-Volhard 1992), and segment identity specification is accomplished in these segments by the Hox genes (McGinnis and Krumlauf 1992). The mandibular development integrates inputs from both the head and the trunk patterning systems (Cohen and Jürgens 1990; Grossniklaus et al. 1994; Vincent et al. 1997). Anterior to the mandible, however, no pair-rule patterning is observed and the anterior-most expression of a gene of the Hox cluster is in the intercalary segment (Abzhanov and Kaufman 1999; Bucher and Wimmer 2005). For patterning of anterior head segments, Drosophila makes use of the so-called head gap-like genes orthodenticle (otd), empty spiracles (ems), and buttonhead (btd). They are expressed early in embryogenesis in broad and overlapping domains (Wimmer et al. 1997). Also their mutant phenotypes are overlapping and correspond roughly to the expression patterns (Cohen and Jürgens 1990; Cohen and Jürgens 1991). However, the functional analysis of the orthologs of these head gap-like genes in another insect — the red flour beetle Tribolium castaneum — revealed that only the late otd function seems similar, whereas ems and btd are not functionally conserved (Schinko et al. 2008). Moreover, another gap gene — knirps (kni) — which in Drosophila is not required for head segmentation is necessary for the development of the antennal and mandibular segments in Tribolium (Cerny et al. 2008).
The detected differences in gene function between Drosophila and Tribolium reflect a variation between insect species in the use of genes at the level of the first zygotically active genes, the gap genes. This might reflect the fact that early development of related species is more variable than development shortly after gastrulation. The stage of greatest similarity between the members of a phylum was termed “phylotypic stage” by Sander (1983). Earlier developmental stages are highly variable due to adaptations to particular modes of reproduction, and thereafter, development diverges toward the differently specialized postembryonic stages, which are again susceptible to adaptive changes. However, the phylotypic stage is not only by morphological criteria the most conserved between the different members of each phylum but represents also the stage with the most conserved gene expression patterns. The recognition that segment polarity genes like engrailed (en), which define the borders of segments in all arthropods analyzed to date, are expressed at this phylotypic stage indicates that the metamerization process of the arthropods is at least partially conserved (Patel 1994; Damen 2002; Chipman and Akam 2008). Very recently, the functional involvement of the segment polarity gene hedgehog (hh) has even been shown in the segmentation of an annelid (Dray et al. 2010) which indicates a common origin of metamerization for all protostomes. Also the metamerization of the arthropod head segments involves segment polarity genes (Rogers and Kaufman 1997; Pechmann et al. 2009); however, the exact mechanisms on how the head gap-like genes direct the metameric expression of the segment polarity genes in the anterior head is still unknown.
About the functional role of the segment polarity genes — e.g., en, hh, and wingless (wg) — in anterior head development, little is known despite the facts that they are compellingly conserved across the arthropods, and their expression patterns have been widely used to mark head segments. There is also not much known about the initial activation of the segment polarity genes in the anterior insect head region — where pair-rule genes are not functional. Moreover, in Drosophila, it has been shown that the cross-regulatory interactions among segment polarity genes in the anterior head region differ from the trunk and have been reported to be specific for each of the anterior head segments — ocular, antennal, and intercalary — while their interaction is identical in the mandibular and all posterior segments (Gallitano-Mendel and Finkelstein 1997). This also suggests a unique establishment of each of the anterior head segments in contrast to a common generation of the posterior segments. Interestingly, these different modes of regulation by segment polarity genes in anterior and posterior head segments may be due to their independent evolutionary origin. Classical embryology has revealed that the subdivision of the coelom — one important feature of segmentation — occurs in two different ways. Anterior segments arise by concomitant subdivision of one large coelom, giving rise to so-called primary or larval segments. Coelomic sacs of the more posterior segments, by contrast, are usually formed one by one from a posterior growth zone (Remane 1950). These latter segments are known as secondary segments. Such a bimodal segmentation is easily observed, for example, in some crustaceans: The nauplius larva typically forms three larval segments, namely first and second antennal segment (the latter corresponds to the intercalary/tritocerebral segment in insects) and the mandibular segment (Scholtz 2000). In this respect, the procephalic segments and the mandibular segment are correlates of larval segments, while the remaining gnathocephalic and trunk segments are of the post-larval type. Differences in the regulation of segment polarity genes between anterior and posterior segments in the insect head could therefore reflect this ancestral subdivision in primary and secondary segmentation (Minelli 2001; Tautz 2004).
In order to identify novel components of the gene regulatory networks that govern anterior head metamerization, we started a bottom-up approach by detecting and functional dissecting cis-regulatory regions of the earliest expressed segment polarity genes, wg and hh, in the anterior head region (Mohler 1995). Such an approach can lead to the identification of transcription factors directly involved in the gene networks patterning each of the anterior head segments. Here we present a functional dissection of wg and hh cis-regulatory regions which led to the isolation of the intercalary-specific enhancer element, ic-CRE. The functional isolation of such a segment-specific cis-regulatory element supports the theory of a unique mode of establishment of each of the procephalic head segments. Moreover, we provide evidence that the establishment of the intercalary segment is delayed in comparison to the other head segments as has been described for Tribolium (Posnien and Bucher 2010; Schaeper et al. 2010).
Materials and methods
Determination of hh transcription start site
The transcription start site of hh was determined at −353 relative to ATG by 5′ RACE PCR on 0–12 h embryonic cDNA 5′ RACE pool using the primer TTGGAGCTGGAACTGGAACTGGAACTG. mRNA from 0 to 12 h embryos was initially isolated using oligo(dT)-coated magnetic beads (Roche, Mannheim, Germany). The cDNA 5′ RACE pool was synthesized using SMART PCR RACE cDNA Synthesis Kit (ClonTech, Heidelberg, Germany).
Reporter constructs
turboGFP reporter [tgfp_SV40 952 bp sequence] was excised with AgeI (site T4 blunted)/AflII from pTGFP_PRL (EVROGEN, Moscow, Russia) and inserted into PmlI (T4 blunted)/AflII of pSLaf1180af vector (Horn and Wimmer 2000) to generate pSLaf_tgfp_af.2. The promoter sequence of hh (−120_+99 bp) was isolated with primers CAACGCGGAATGAACTCGAGGCGATAG (XhoI_Forward) and AACTAGTTAGCTCTCGGTTCGGACAACCGTTG (SpeI_Reverse) on Drosophila genomic DNA and subcloned into Xho/SpeI of pslaf_tgfp_af.2 to result in construct pSLaf[Dm_hh promoter_tgfp_SV40]. The promoter sequence of wg (−159_+121 bp around tsA) was isolated with primers CTCGAGCAGGAGTCAGGGTATAGCTCCAC (XhoI_Forward) and ACTAGTTTCGATAGAATACACTCGGCTCGCTCTAG (SpeI_Reverse) and subcloned into Xho/SpeI of pSLaf_tgfp_af.2 to result in construct pSLaf[Dm_wg promoter_tgfp_SV40]. The 156 bp hs43 promoter sequence was excised from pSLaf_hs43_lacZ_af with XhoI/PstI (T4 blunted) and subcloned into XhoI/SpeI (T4 blunted) of pSLaf_tgfp_af.2 to result in construct pSLaf[hs43_tgfp_SV40]. pSLaf_hs43_lacZ_af was constructed by excising (HindIII/XhoI) a 4.4 kb [hs43_lacZ_SV40] fragment from pCasper_hs43_lacZ (Thummel and Pirotta 1992) and ligating it into pSLaf1180af vector (Horn and Wimmer 2000). The DNA sequence spanning −6902_+265 bp of hh locus was amplified on genomic DNA template by long-range PCR (High Fidelity Enzyme, Fermentas, St. Leon-Rot, Germany) with primers CGAGCAGCATTGTGAGGGAGCACACTACA, forward and GCACTTCACTTTTGGCACACAGACACGCT, reverse and cloned via T/A ligation in the PCRII vector (Invitrogen, Darmstadt, Germany) to result in pTAII_Dm_hh_upstream(7.16). DNA sequence spanning −8,094 bp upstream of wg tsA to +193 bp downstream of tsB (i.e., +2,122 bp downstream of tsA) was amplified with primers CTCGACGGCAAACAGAGAAGGCGAGGAGTGACT, forward and AGTCACTCCTCGCCTTCTCTGTTTGCCGTCGAG, reverse and cloned in PCRII to result in pTAII_Dm_wg_ upstream(−8.1). Sequence spanning −16,212_−7,813 kb upstream of wg tsA was amplified with primers GCTGCTCCAGATCATCAGCGTTGTACCAG, forward and GAATCGGAATCGGGTTGGCTCGACCTCAC, reverse and cloned in PCRII to result in pTAII_Dm_wg_ upstream(−16.2_−7.8). The hh and wg upstream sequences were excised with NsiI (−6.43 kb)_NotI (PCRII polylinker) from pTAII_Dm_hh_ upstream(7.16) and with EcoRI from pTAII_Dm_wg_ upstream(−8.1) and subcloned into the polylinker of pSLaf_lacZ_af to generate pSLaf[−6.43 kb_hh upstream_lacZ_SV40] and pSLa[−8.1 kb_wg upstream_lacZ_SV40], respectively. pSLaf_lacZ_af was constructed by partially digesting the pSLaf_hs43_lacZ_af with PstI to remove the 197 bp hs43 fragment followed by autoligation. Also the −8.1_−3.9 and −6.7_−3.8 kb hh upstream subfragments (resulting from EcoRI/HpaI and XhoI restriction, respectively) were subcloned into pSLaf_hs43_lacZ_af to generate pSLaf[−8.1_−3.9 kb hh upstream_hs43_lacZ_SV40] and pSLaf[−6.7_−3.8 kb hh upstream_hs43_lacZ_SV40], respectively. The −16,212_−7,813 kb (wg tsA) sequence was excised (KpnI_NotI) from pTAII_Dm_wg_ upstream(−16.2_−7.8) and subcloned into pSLaf[Dm_wg promoter_tgfp_SV40] to generate pSLaf[−16.2_−7.8 wg upstream_wg promoter_tgfp_SV40]. The 1,009 bp α fragment was isolated with primers TCGCGAGCTGATAGCACAATGGACCCAC, forward and CTCGAGTATCTAAAAGCCAATTTCGATTGTGAC, reverse and cloned into pSLaf[Dm_hh promoter_tgfp_SV40] and pSLaf[hs43_tgfp_SV40] to generate pSLaf[α_hh promoter_tgfp_SV40] and pSLaf[α_hs43_tgfp_SV40], respectively. Similarly, the overlapping subfragments (γ1, β4, β3, γ2, F6_R5, F3_R2, F5_R4) were isolated with proofreading PCR (primers are available upon request; Ntini 2009) and cloned in pSLaf[Dm_hh promoter_tgfp_SV40] to generate the respective pSLaf[γ1/β4/β3/γ2/F6_R5/F3_R2/F5_R4_hh promoter_tgfp_SV40] constructs. γ1 and F5_R4 were also cloned into pSLaf[hs43_tgfp_SV40] to generate pSLaf[γ1_hs43_tgfp_SV40] and pSLaf[F5_R4_hs43_tgfp_SV40], respectively. Cassettes consisting of [cis-regulatory region_promoter_tGFP_SV40] or [cis-regulatory region_promoter_lacZ_SV40] were finally excised with AscI from the above pSLaf_constructs and subcloned into pBac[3xP3_EGFPafm] (Horn and Wimmer 2000) to generate the respective piggyBac constructs. The ic-CRE γ1mF3-γ1mF6 subfragments (resulting from 5′ fragmentation of the γ1; primers are available upon request; Ntini 2009) were subcloned in pSLaf[Dm_hh promoter_tgfp_SV40] resulting into pSLaf[γ1mF3/F4/F5/F6_hh promoter_tgfp_SV40] and finally integrated via attB-attP site-specific recombination.
Transgenesis
For piggyBac-mediated transgenesis, the generated piggyBac constructs (please see “Reporter constructs”) were coinjected at 500–1,200 ng/μl (we observed that increasing the pBac construct concentration enhanced transformation efficiency of large constructs) with helper plasmid providing transposase activity (phspBac) at 300 ng/μl (Horn et al. 2000). For the piggyBac random insertions, at least two independent lines were analyzed to circumvent the possibility of position effects. For site-specific transgenesis using the attB-attP φC31-mediated integration system (Bischof et al. 2007), the complete 314 bp attB sequence was excised from pTA-attB (Calos MP, Stanford University, personal communication) with EcoRI; restricted ends were blunted with T4 DNA polymerase and subcloned into pBac[3xP3_EGFPafm] (Horn and Wimmer 2000) linearized with BglII (T4 blunted) generating the vector pBac_attB. The γ1mF3-γ1mF6 reporter cassettes were excised with AscI from pSLaf[γ1mF3/F4/F5/F6_hh promoter_tgfp_SV40] and subcloned in the pBac_attB vector to generate the respective pB_attB constructs (for example, pB_attB[γ1mF5_hh promoter_tgfp_SV40]). All the generated pB_attB constructs were injected and successfully assayed in the line bearing the attP landing site at position 96E of the third chromosome (Ac. Num EF362408). This is a combined line carrying on the X chromosome a codon-optimized φC31 integrase driven under the control of the vasa promoter (Bischof et al. 2007).
Whole mount embryo in situ hybridization
To generate Dig- or Fluo-labeled RNA probes for in situ hybridization, cDNA sequences of genes of interest were cloned in the PCRII vector (Invitrogen, Darmstadt, Germany) and antisense-RNA was generated from T7 or Sp6 promoter using the respective RNA polymerase (Roche, Mannheim, Germany). In vitro transcription was in the presence of 10% DIG-labeling or Fluo-labeling rNTP mix (Roche, Mannheim, Germany); 0–10.5-h embryo collections were dechorionated for 3 min in 50% chlorix and fixed in 2 ml heptane, 3.7% formaldehyde in 1.5 ml PEM (0.1 M PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.9) for 20 min at rt. Double in situ hybridization was as in Rehm et al. (2009), apart from an additional 30-min detergent treatment (1% SDS, 0.5% Tween-20, 50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl) before prehybridization. NBT/BCIP staining was in AP buffer, pH 9.5, and FastRed (Sigma-Aldrich, Munich, Germany) staining in AP buffer, pH 8.2.
Microscopy
Embryos stained after in situ hybridization were mounted in glycerol (~90%) and documented with a Zeiss Axioplan 2 microscope (×20 or ×40 planes) using the ImageProPlus software (Version 6.2; MediaCybernetics). Embryonic staging was after Campos-Ortega and Hartenstein (1997).
In silico analysis of DNA sequences
In silico analysis to identify putative transcription factor binding sites within cis-regulatory regions was performed using the MatInspector (Cartharius et al. 2005; http://www.genomatix.de/online_help/help_matinspector/matinspector_help.html; http://www.genomatix.de/cgi-bin/matinspector_prof/mat_fam.pl). Predictions were inspected manually, checked in correlation with literature reports, and filtered through phylogenetic conservation using the Drosophila EvoPrinter (http://evoprinter.ninds.nih.gov/evoprintprogramHD/evphd.html) or the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
Results and discussion
A distinct cis-regulatory element controls wg expression during foregut development at the anterior terminal pole
A cis-regulatory region, WLZ4.5L, spanning 4.8 kb upstream of the wg transcription start site A (tsA) had been previously assayed by Lessing and Nusse (1998). Their construct mediates reporter expression in the wg gnathal and trunk stripes from the early onset of the wg expression at the blastoderm stage and then during germ band extension stages but lacks the metameric expression pattern in the procephalon. In search for anterior head segment-specific cis-regulatory elements, we thus assayed two overlapping 5′-extended DNA fragments (−6.7_−3.8 kb and −8.1_−3.9 kb) combined with an hs43 basal promoter (Thummel and Pirotta 1992; Fig. 1A). The larger fragment (−8.1_−3.9 kb) mediates reporter expression in the anterior-most terminal region at the cellular blastoderm stage (Fig. 1H, I) overlapping the endogenous anterior-most wg expression domain (Fig. 1E, F). During germ band extension the reporter is expressed in the foregut anlage but not in the labral spot (Fig. 1J). Because the smaller fragment (−6.7_−3.8 kb) does not mediate expression in this region (Fig. 1G), we conclude that the region between −8.1 and −6.7 kb contains cis-regulatory elements essential for transcriptional control of wg foregut expression. Please note that the −6.7_−3.8 kb fragment mediates “patched” reporter expression in the trunk stripes of stage 11 embryos, excluding the gnathal segments. This pattern seems wg-specific as it overlaps with endogenous wg expression and turns on when the WLZ4.5L-mediated expression fades (Lessing and Nusse 1998). The fact that the larger fragment (−8.1_−3.9 kb) does not report this expression indicates a rather complex regulation during the maintenance phase of wg expression.
Fig. 1.
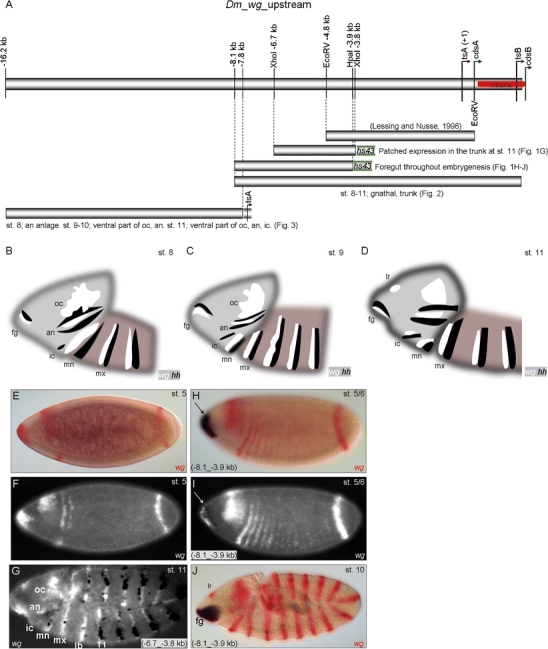
Functional dissection of the wg upstream region. a Schematic representation of partial wg locus and the fragments assayed for enhancer function in vivo. Bars represent the −4.8-kb fragment assayed by Lessing and Nusse (1998), the (−6.7_−3.8-kb) and (−8.1_−3.9-kb) fragments assayed in combination with hs43 basal promoter, the 10.216-kb fragment (spanning 8,094 bp upstream of tsA to +195 bp downstream of tsB), and the 8.4-kb fragment (−16.2_7.8 kb) assayed in combination with the endogenous tsA promoter (−159_+121 bp). b–d Schematic representation of wg (white) and hh (black) expression at different embryonic stages. e–j Double in situ hybridization of wg (FastRed staining or white fluorescent in f, g, i) and the mediated reporter gene expression (NBT/BCIP-blue staining or black in g, i). e, f Endogenous blastodermal wg expression. g Fragment (−6.7_−3.8 kb) mediates expression in cells of the trunk segments, excluding the gnathal segments (mn, mx, lb) at stage 11. h, i Fragment (−8.1_−3.9 kb) mediates expression in the anterior-most terminal region at blastodermal stages overlapping the endogenous wg expression. j During germ band extension, the same enhancer fragment mediates expression in the foregut anlage (fg). oc ocular, an antennal, ic intercalary, mn mandibular, mx maxillary, lb labial, lr labrum
Procephalic and trunk metamerization employ separate cis-regulatory elements controlling wg expression
Lessing and Nusse (1998) used in their WLZ4.5L construct an endogenous wg promoter. To exclude the possibility that the reason we did not find any procephalic regulatory elements is based on the use of a heterologous promoter (hs43 basal element), we made a large construct that contains all the upstream regions so far analyzed plus both previously identified transcriptional promoters (tsA and tsB) of wg spanning the region −8.094 kb upstream of tsA to +195 bp downstream of tsB (NCBI reference sequences NM_078778.3 and NM_164746.1, respectively). This 10.216 kb fragment mediates reporter expression in the gnathal and trunk stripes, as well as in the labral spot, the foregut region, and in excess in the prospective hind gut region (similar to WLZ4.5L; Lessing and Nusse 1998), but still it lacks cis-regulatory information for the procephalic stripes (Fig. 2).
Fig. 2.
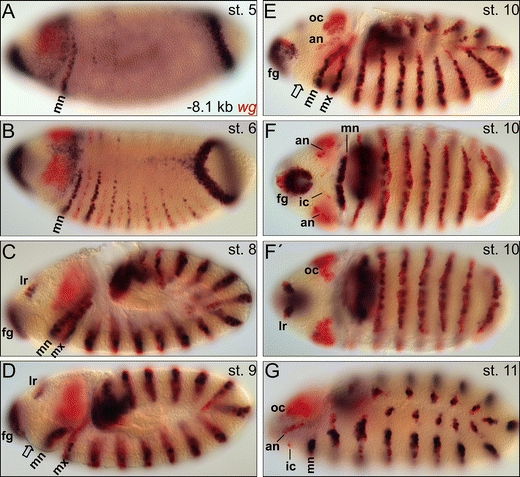
Expression pattern mediated by the −8.1 kb upstream region of wg (10.216 kb fragment). A, B At stages 5 and 6, the mediated reporter expression pattern in the even-numbered stripes comes up with a delay compared to the endogenous wg expression. C–G During germ band extension, the −8.1 kb region mediates the complete expression pattern in the trunk segments, foregut (fg), and labrum (lr) but not in the procephalon. The open arrow in D and E depicts the region of the prospective intercalary segment. For abbreviations, see Fig. 1
At the late blastoderm stage 5/6, the even-numbered reporter stripes come up delayed compared to the endogenous wg stripes (Fig. 2A, B). Again this is reminiscent of the expression pattern generated by WLZ4.5L (Lessing and Nusse 1998). On the contrary however, the reporter expression generated by that construct fades from the epidermis of late germ bands (stage 11) whereas the reporter stripes generated by the 10.216 kb fragment follow the endogenous trunk wg expression pattern splitting into ventral and dorsal subdomains within each segment (Fig. 2G). Thus, in contrast to WLZ4.5L, the 10.216 kb fragment contains additional cis-regulatory elements required for the proper maintenance mode of wg expression in the trunk. Nevertheless, anterior head segment-specific cis-regulatory elements are not included in this 10.2 kb cis-regulatory region of wg which sufficiently mediates expression in the trunk and the anterior-most terminal region during germ band extension. This indicates that cis-regulatory information governing wg expression in the procephalic head segments must be distinctive, which supports the idea that the processes of primary and secondary segmentation employ distinct transcriptional regulatory networks.
In this respect, the anterior-most segment that is ruled by the secondary “trunk” segmentation mode is the mandibular segment, while the wg expression in the three procephalic segments must be set up in an independent manner (Fig. 2E–G). This divide into different segmentation modes seems to be at a similar position as in Tribolium, for which it has been shown that pair-rule gene mutants or knock downs affect the mandibular and more posterior segments (Posnien and Bucher 2010; Maderspacher et al. 1998; Choe et al. 2006; Choe and Brown 2007, 2009). Thus, despite the difference between long — Drosophila — versus short — Tribolium — germ band mode (Wolff et al. 1995), there seems to be a common point for the phase shift between anterior (primary) and posterior (secondary) segmentation.
Upstream sequence of the wg gene locus contains different types of procephalic cis-regulatory information for establishment and maintenance modes of expression
In the search for cis-regulatory elements governing expression of wg in the procephalic region, 8.4 kb of further upstream sequence was isolated, spanning region −16.212 to −7.813 kb relative to tsA and assayed in combination with a 280 bp endogenous promoter region surrounding tsA (−159 to +121 bp; Fig. 1A). This upstream region also mediates the metameric expression pattern of wg; however, the reporter trunk stripes do not come up at the early cellular blastoderm stage 5 but only at stage 6/7 (Fig. 3A). The reporter expression thus lacks the early onset of trunk expression at blastoderm stage as it is mediated by the −8.1 kb enhancer. In addition, this further upstream enhancer fragment mediates reporter expression in the cell stripe of the antennal segment primordium at stage 7/8 (Fig. 3B, C). This is the time point when the endogenous wg antennal expression domain first appears at the lateral procephalic ectoderm (Liu et al. 2006). Since the first detection of the antennal-specific reporter expression pattern coincides with formation of the endogenous wg antennal stripe, the −16.212- to −7.813 kb enhancer fragment probably contains cis-regulatory elements underlying the establishment of procephalic wg expression specific for the antennal segment. In contrast, the mediated reporter expression pattern does not overlap the anterior procephalic expression domain of wg at the blastoderm stage corresponding to the presumptive ocular region, the so-called head blob (Liu et al. 2006). Actually the first reporter expression in this ocular region is detectable only at stage 8 in the ventral-most cells of the head blob (Fig. 3C). Also during germ band extension, reporter expression is mediated only in the ventral-most part of the ocular region (Fig. 3D–G). The mediated reporter expression pattern in the intercalary segment comes up later, at stage 11, overlapping the endogenous wg intercalary spot (Fig. 3G) which is already detected clearly during stage 10 (Figs. 2f and 3F, F′; Gallitano-Mendel and Finkelstein 1997; Liu et al. 2006). The −16.212- to −7.813 kb enhancer fragment, therefore, does not mediate the onset of the endogenous wg expression in the intercalary segment.
Fig. 3.
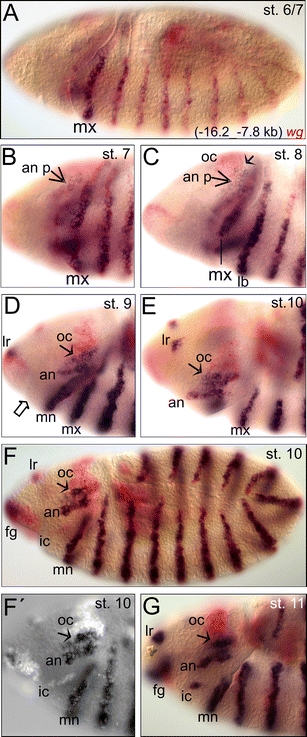
Expression pattern mediated by the (−16.2_−7.8 kb) fragment of wg. A The earliest mediated reporter expression pattern is at stages 6/7 in the trunk stripes. B, C At stages 7/8, the enhancer fragment mediates expression also in cells of the antennal primordium (an p, arrow). This is the point when the endogenous wg antennal stripe emerges at the lateral procephalic ectoderm. The short arrow in C depicts ventral-most cells within the ocular region that express the reporter at stage 8. D–F, F′ At stages 9 and 10, the enhancer mediates expression in the antennal stripe and the ventral-most part of the ocular segment (arrow) but not in the intercalary segment. The open arrow in D depicts the region of the prospective intercalary segment. G The mediated reporter expression in the intercalary segment appears at stage 11 overlapping the endogenous wg intercalary spot. For abbreviations, see Fig. 1
This delay in the reporter expression implies that the set up of the intercalary-specific wg expression is under the control of separate cis-regulatory information and reflects that regulation of segment polarity gene expression can be divided into sequential phases of establishment and maintenance. The cis-regulatory elements mediating the intercalary expression included in this upstream enhancer seem to be rather involved in the maintenance of wg expression. This is consistent with the fact that also for the trunk segments this further upstream region mediates the maintenance phase of wg expression rather than the onset (Fig. 3A, G). Only for the antennal expression, this further upstream element seems to contain all cis-regulatory information for both establishment as well as maintenance of wg expression (Fig. 3B–G).
Transcriptional control of wg in the ocular region is mediated by distinct dorso- and ventral-specific cis-regulatory elements
In principle, the anterior-most procephalic expression domain of wg corresponds to a proposed segmental unit, i.e., the ocular segment (Schmidt-Ott and Technau 1992), based on data of the phylogenetically conserved expression pattern of engrailed (Schmidt-Ott et al. 1994; Urbach and Technau 2003). In more primitive insects, it splits into expression subdomains, namely the median protocerebral neuroectoderm expression domain (mpn), the dorsal protocerebral (dpn), and the ventral protocerebral neuroectoderm domain (vpn; Liu et al. 2006). On the contrary, in Drosophila, it remains intact constituting the “head blob” (Schmidt-Ott and Technau 1992). The vpn domain has been specifically lost in Drosophila, and as reported in Liu et al. (2006), the contiguous protocerebral neuroectoderm domain (pne) or head blob may be equivalent to (a) the mpn, (b) the dpn, or (c) the primordial yet non-dissociated protocerebral ectoderm domain of primitive insects. The 8.4 kb wg upstream sequence (−16.2 to −7.8 kb) contains cis-regulatory information that drives expression only in the ventral part of the head blob (Fig. 3D–G). These data indicate distinct dorsoventral cis-regulatory information controlling expression of wg within the ocular segment and support the idea that the ventral-specific expression subdomain of the contiguous wg head blob most likely corresponds to the median protocerebral expression domain of less derived insects, with respect to its topological orientation.
Dorsoventral differences in the regulation of segment polarity genes have also been reported in the context of the anterior head segment-specific cross-regulatory networks. For instance, wg represses hh expression in the dorsal part of the ocular and antennal segments, while it maintains hh expression ventrally within the same segments (Gallitano-Mendel and Finkelstein 1997). Thus, transcriptional control of segment polarity genes in the procephalic region may involve dorso- and ventral-specific cis-regulatory elements functional within the very same segmental unit. In a developmental context, this may reflect the response of segment polarity genes to distinct signals in different parts of the ocular and antennal segments.
Identification of an intercalary-specific cis-regulatory element of hh
The transcription start site (tss) of hh was identified by 5′ RACE PCR to be located −353 bp relative to translation start site, which is different from the tss reported in Lee et al. (1992), at −385 bp. This may be due to a nucleotide polymorphism (T > C) that the strain we used carries at position −387. Moreover, the annotated EST, EK111112.5prime, starts at position −374 bp and is also affected by a polymorphism (C > G) right at this position. For the following constructs, the numbering refers to the transcription start site we identified being +1.
A 6.43 kb upstream sequence plus endogenous promoter and part of the 5′ UTR (−6.43 kb to +265 bp) were initially assayed (Fig. 4A–I). At blastoderm stage, this upstream sequence mediates expression of the reporter in an anterior domain broadly overlapping the early endogenous procephalic expression domain of hh (Mohler 1995; Fig. 4B, C), while it is not mediating any expression in the presumptive trunk. The odd-numbered reporter stripes appear at stage 8 (Fig. 4E) followed up with a delay by the even-numbered ones (Fig. 4F, G). During germ band extension, the reporter is also expressed in the procephalic head stripes (Fig. 4G–I). This 6.43 kb upstream region was then dissected into 5′ shortened fragments (Fig. 4A). The −4.08 kb fragment mediates expression in the intercalary segment and in some dorsal epidermal cells (Fig. 4J–L), while the −3.17 kb fragment does not retain expression in the intercalary segment (Fig. 4M). Therefore, the region between −4.08 and −3.17 kb (represented by the red bar in Fig. 4A) must contain cis-regulatory information essential for the transcriptional control of hh expression in the intercalary segment.
Fig. 4.
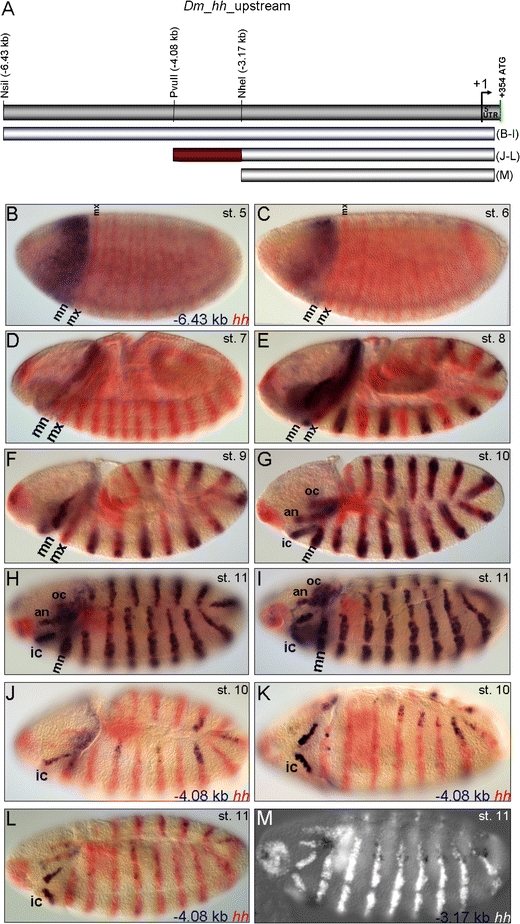
Functional dissection of the hh upstream region. A Schematic representation of the fragments assayed for enhancer activity. B, C The −6.43 kb fragment mediates expression in an anterior domain broadly overlapping the endogenous anterior expression domain of hh at blastoderm stage. D–F The even-numbered stripes of the mediated reporter expression pattern come up after the odd-numbered ones have fully developed. G–I By the completion of germ band extension, the enhancer mediates expression in the trunk and procephalic stripes. J, K The −4.08 kb fragment mediates expression in the intercalary segment while M the −3.17 kb fragment does not. Thus, the −4.08_−3.17 kb region (red bar in A) is an essential element for intercalary-specific transcriptional control of hh termed ic-CRE. For abbreviations, see Fig. 1
To test whether this enhancer fragment is also sufficient to ensure intercalary-specific expression of hh, the sequence from −4.08 to −3.077 kb (named thereafter α fragment; Fig. 5A) was assayed in combination with an hs43 basal promoter (Thummel and Pirotta 1992) or with the endogenous hh promoter region (−120 to +99 bp). Expression of the reporter was specifically mediated in the intercalary segment (plus a few cells in the mandibular and maxillary segments) when the endogenous promoter was used (Fig. 5B–D) but not in combination with the heterologous promoter (data not shown). Therefore, the α fragment is essential and sufficient for the transcriptional control of the intercalary-specific expression of hh. This sequence and its functional subfragments are thus referred to as the intercalary-specific cis-regulatory element (ic-CRE). In an effort to further restrict the cis-regulatory element crucial for intercalary-specific transcriptional control of hh, we first performed a phylogenetic conservation analysis (Bejerano et al. 2005; see “Materials and methods”) of the ic-CRE sequence to detect highly conserved sequence blocks (Fig. 5A; EN and EAW unpublished). Second, the 1 kb ic-CRE sequence (α fragment) was further dissected by assaying overlapping subfragments or 5′ truncated sequences that end at a common point, −3,465 bp (Fig. 5A), together with the endogenous hh promoter. During this dissection analysis, fragments were designed so that highly conserved sequence blocks were not disrupted. The 5′ truncation of the γ1 sequence was assayed at the same genomic integration site using the attP–attB site-specific recombination system (Bischof et al. 2007). This system was selected to overcome potential position effects by random integration resulting from piggyBac-based germ line transformation (Horn et al. 2000). For that reason, we examined at least two independent transgenic lines when using the piggyBac-mediated transformation system. The 335 bp sequence F5_R4 (−3,799_−3,465 bp; Fig. 5A) was the minimum sequence assayed that still retains expression in the intercalary segment with an onset of expression at stage 10 (Fig. 5E–H) and with a partial and spotty metameric expression later in the trunk (Fig. 5H). In contrast, the 450 bp ic-CRE sequence termed γ1mF5 (−3,914_−3,465 bp) covering all of and extending the 335 bp fragment (Fig. 5A) mediates expression specific to the intercalary segment already from stage 9 on (Fig. 5I–L). This fragment, however, still lacks the very early onset of endogenous hh expression in the intercalary segment anlage at stage 8, which is mediated by the 621 bp ic-CRE fragment γ1 (arrow in Fig. 5M, N) and the γ1mF3 subfragment (Fig. 6A, C). To further confirm the requirement of the ic-CRE to interact with the endogenous hh promoter, the 621 bp γ1 and the 335 bp F5_R4 subfragments were also assayed in combination with the hs43 basal promoter, again not showing the ic-CRE-mediated expression.
Fig. 5.
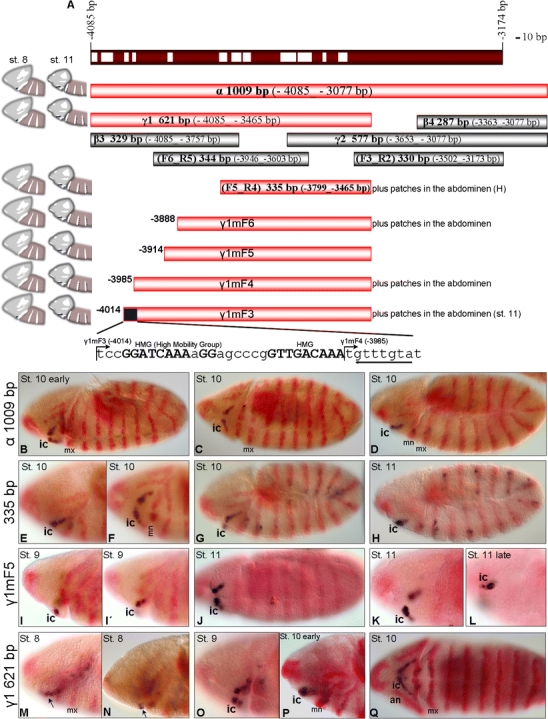
Functional dissection of the ic-CRE. A Schematic representation of the analyzed ic-CRE fragments and the mediated reporter expression pattern (dark blue) at stages 8 and 11 (in reference to Fig. 1b, d). White bars within the red (−4,085 to −3,174 bp) fragment (top of the panel) represent 12 Drosophilidae phylogenetic conservation at sequence level. Fragments that—in combination with the endogenous hh promoter—mediate expression in the intercalary (ic) segment are labeled light red. The blue box (bottom of the panel) constitutes a cluster of in silico predicted HMG sites. Capitals represent 12 Drosophila species conservation. Underlined is a putative HMG site found in the reverse complement orientation displaying 11 species conservation. B–D The fragment “α” mediates specific expression in the ic segment and in a few cells in the mn and mx segments. E–H The 335-bp fragment (F5_R4) is partially de-repressed from late stage 10 on in the trunk. I–L The γ1mF5 fragment mediates specific expression in the ic segment. M–Q The γ1 fragment mediates the early onset of the reporter expression in the intercalary segment anlage at stage 8 (arrow in M, N), which is also mediated by the γ1mF3 fragment (see Fig. 6A, C) but not by γ1mF4. This indicates that the 5′ part of γ1mF3 (blue box in A) contains cis-regulatory elements necessary for the early onset of hh expression in the ic segment. For abbreviations, see Fig. 1
Fig. 6.
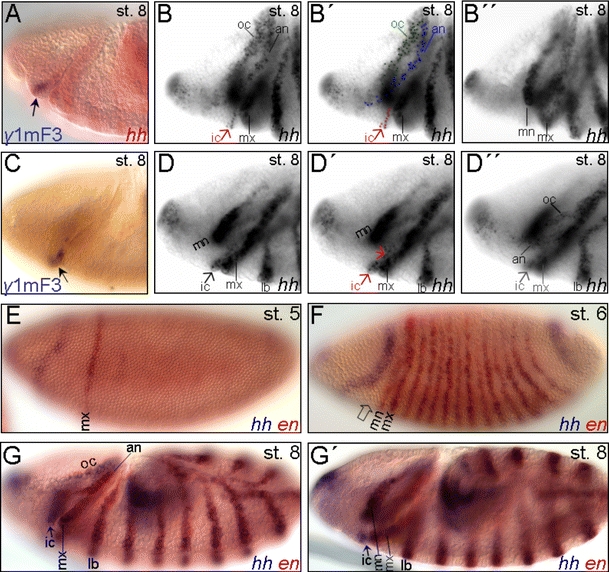
Early onset of the intercalary-specific expression of hh. A, C The γ1mF3 enhancer fragment (see Fig. 5A) mediates the early onset of reporter expression in the intercalary segment anlage (arrow) at stage 8 (see also Fig. 5M, N). B, B′′, D, D′′ Early procephalic expression of hh at stage 8. B, B′′ The procephalic stripes (oc, an, ic) are detected at a different focal plane (B, B′) than the mn stripe (B′′). B, B′′ In this lateral view, although detected at the same focal plane, the cell stripe in the intercalary (ic) segment anlage (arrow) is discontinuous from the antennal (an) segment anlage which progressively delineates from the ocular (oc) one. D, D′′ In this ventrolateral view, the cell stripes in the an and oc segment anlagen are detected at the same focal plane (D′′) while the ic is out of focus and vice versa. D, D' The slightly different focal plane of D′ compared to D allows the cell groups of both the ic anlagen to be detected. E–G′ Double in situ hybridization of hh (purple) and en (red). F Anterior to the mn stripe, the early procephalic expression domain of hh progressively splits into the antennal and ocular primordium during stage 7. The cells at the posterior margin of the early procephalic hh domain co-expressing en are precursors of cells of the antennal ectodermal stripe formed at the posterior procephalic margin at stage 8 (G). The open arrow depicts the precursor cells of the presumptive ic segment anlage. G, G′ Different focal planes of the same embryo at stage 8. hh but not en expression is detected in the ic segment anlage at stage 8 (arrow). For abbreviations, see Fig. 1
The ic-CRE requires promoter-specific interaction
During the functional dissection analysis, we observed that the 1 kb ic-CRE (α fragment) and its functional subfragments mediate the intercalary-specific expression pattern only in combination with the endogenous hh promoter region (−120 to +99 bp) but not with the hs43 TATA-box basal promoter (Thummel and Pirotta 1992). On the other hand, subfragments of the −8.1 kb upstream region of wg mediate specific reporter expression patterns when combined with the same hs43 basal promoter (Fig. 1A, G–J). These results indicate that an enhancer–promoter-specific interaction underlies transcriptional control of the intercalary-specific expression of hh or that the hs43 promoter lacks core promoter elements required to mediate the ic-CRE function. The endogenous hh promoter is TATA-less and contains instead a downstream promoter element (DPE; Butler and Kadonaga 2001; Lim et al. 2004) as one of its core elements. On the other hand, the wg endogenous promoter is also TATA-less but remarkably there is no detectable DPE sequence matching the consensus RGWYV(T). The only detected core promoter element in the wg promoter sequence is the initiator element (consensus TCAKTY; Lim et al. 2004) in the case of tsA, which typically encompasses the transcription start site (the underlined A is +1). Enhancer–promoter specificity has been reported in several cases of transcriptional regulation and may depend on the activity of sequence-specific transcription factors which function as DPE-specific activators (Hsu et al. 2008; Juven-Gershon et al. 2008; Juven-Gershon and Kadonaga 2010). Interestingly, occurrence of the DPE motif in the Drosophila endogenous core-promoters is as common as the TATA-box (Kutach and Kadonaga 2000). Consistently, the reporter expression pattern mediated by the complete −6.43 kb hh upstream region appeared faded when the DPE was disrupted by a point mutation (data not shown), supporting a functional role for DPE activity in embryonic transcriptional control of hh expression.
Early onset of the ic-CRE-mediated expression pattern is ensured by a 30 bp sequence which possibly recruits HMG DNA-binding activity
During the 5′ fragmentation assay of the 621 bp γ1 fragment (Fig. 5A), we identified a 30 bp sequence from −4,014 to −3,985 bp which is required to ensure the early onset of the ic-CRE-mediated expression pattern in the intercalary segment anlage at stage 8, as this expression is mediated (Fig. 6A, C) by the fragment γ1mF3 (−4,014 to −3,465 bp) but not by the fragment γ1mF4 (−3,985 to −3,465 bp). This sequence does not mediate early expression by itself but only in concert with sequences within the 335 bp element, since even the β3 fragment (−4,085 to −3,757 bp) does not mediate reporter gene expression. In silico analysis indicates that this short sequence consists of two highly conserved blocks GGAT C AAAaGG and GTTGA C AAAt separated by a 6 bp stretch (Fig. 5A; capitals represent 12 Drosophila species phylogenetic conservation). Both sequences resemble the binding motif of high mobility group (HMG) protein factors [WCAAAS] (entry in the CDD Database of NCBI: cd01388 “SOX-TCF_HMG-box”; Love et al. 1995; Werner et al. 1995). In addition, they both conform to the consensus binding sequence of HMG-box proteins of the SOX family [WWCAAW] (Churchill et al. 1995; Lefebvre et al. 2007). In silico prediction in the 50 bp DNA sequence (−4,019 to −3,970 bp) using the MatInspector generates a hit in the first block GGA TC AAAaGG, scoring the binding matrix of dTCF (Drosophila T-cell factor homolog or Pangolin) which is WTC AAAS (underlined are the four nucleotides of the core sequence used by MatInspector; Lee and Frasch 2000). The non-conserved A nucleotide which disturbs the conservation block does not match the matrix at the corresponding position 7 (S) (S stands for “strong nucleotide,” i.e., G/C). Still, the site strongly resembles the consensus binding sequence of dTCF determined by PCR-based binding site selection [GATCAAAGG] (van de Wetering et al. 1997) which matches well the canonical Lef1/TCF binding motif [WWTCAAAGG] (van de Wetering et al. 1991, 1993). Only the first block, but not the second one, scores in silico the binding matrix of dTCF, as it seems that a T residue in the (second) W position of the general HMG-box consensus WCAAAS (or WWCAAW) is a prerequisite for specific recognition by the HMG-box of dTCF. Remarkably, one more putative HMG binding site (TACAAAC) lies 3′ juxtaposed to the isolated fragment matching the WCAAAS consensus (at position −3,984 to −3,978 bp, reverse complement). This sequence is filtered through 11 species phylogenetic conservation, with the Drosophila yakuba sequence being divergent.
In conclusion, although the enhancer fragment γ1mF5 (−3,914_−3,465 bp) mediates specific expression in the intercalary segment during stages 9–11, early onset at stage 8 is only ensured by an additional fragment (−4,014_−3,985 bp) which provides early temporal control. HMG DNA-binding activity is predicted in silico in that specific DNA sequence, and collectively the enhancer fragment (−4,014_−3,975 bp) consists of three highly conserved sequence blocks, all of which conform to the HMG DNA-binding consensus. The first block also scores with one mismatch the binding matrix of dTCF; the endogenous sequence, however, is not efficiently recognized by dTCF in vitro (not shown). Still, this cluster of three highly conserved HMG putative binding sites may recruit HMG DNA-binding activity in vivo necessary to ensure the early onset of the ic-CRE-mediated expression pattern. The functional, architectural role of HMG proteins within the context of chromatin environment is attributed to their strong DNA-bending properties, thereby facilitating DNA-binding of sequence-specific factors and the assembly and stabilization of transcriptional complexes (Giese et al. 1997; Dragan et al. 2004). HMG activity has been previously implicated in transcriptional control of other early embryonic developmental processes as well (reviewed in Dailey and Basilico 2001). In particular, members of the Sox protein family are expressed temporal and spatiospecific and can interact with other sequence-specific transcription factors to control crucial aspects of developmental gene expression (Kamachi et al. 1998, 2001; Wilson and Koopman 2002; Kondoh and Kamachi 2010). In Drosophila, eight Sox genes have been characterized (Crémazy et al. 2001; McKimmie et al. 2005). Function of the fish-hook/Dichaete/Sox70D protein, containing an HMG domain homologous to that of the mammalian Sox2, was shown essential for segmentation in the early Drosophila embryo (Nambu and Nambu 1996; Russell et al. 1996; Sánchez-Soriano and Russell 2000). In mouse, the temporal-specific late onset of Sox9 expression correlates with the timing of neuron-to-glial switching, being thereby involved in cell-type specification (Stolt et al. 2003). Interestingly, it was recently shown in zebrafish that Sox factors crucially control the timing of biphasic target gene expression, since their activation threshold determines the onset of the second phase of specific target gene expression (Onichtchouk et al. 2010).
Delayed and distinct establishment of the intercalary segment
The functional detection of an early active cis-regulatory control element underlying the onset of the intercalary-specific hh expression led us to address the developmental issue of formation of the intercalary segment. Therefore, we re-examined the mode of establishment of segment polarity gene expression in the intercalary segment anlage in comparison to the rest of the procephalic head segments (Fig. 6). The complex morphogenetic movements during the early gastrulation, marked by the formation of the cephalic furrow, makes it hard to clearly define the primordia of the procephalic segments and ascribe them back to the blastoderm fate map. Lateral embryonic views of stages 7/8 (Fig. 6B, B′) are misleading to assume that the ventrally located intercalary stripe, marked by the expression of hh (from stage 8 on) and en (from stage 10 on), arises from splitting from the more dorsal ectodermal antennal stripe (Gallitano-Mendel and Finkelstein 1997; Mohler 1995; de Velasco et al. 2006). However, from slightly twisted ventrolateral views (Fig. 6D–D′′) it can be seen that the onset of hh expression in the intercalary segment anlagen at stage 8 is detected at a focal plane which is different from detection of the antennal and ocular stripes. This indicates that formation of the intercalary hh stripe arises in a distinct and independent set of embryonic cells that are not in direct continuation to the antennal and ocular hh expressing cells. In addition, at stage 8, the hh antennal and ocular stripes are still in contact at their dorsal- and ventral-most ends (Fig. 6D′′), suggesting that they arise from progressive separation (or splitting) of the early procephalic anterior wide expression domain of hh (Fig. 6F; Chang et al. 2001). The en co-expressing cells at the posterior margin of this domain (at stage 6; Fig. 6F) will subsequently belong to the antennal stripe formed and defined at the posterior margin of the procephalic ectoderm (Fig. 6G; Schmidt-Ott and Technau 1992). Since not only hh (stage 8) but also wg and en (both at stage 10) are expressed late in the intercalary segment compared to the other segments (Schmidt-Ott and Technau 1992), we postulate that the intercalary segment anlage is formed by the ventral ectodermal cells lying anterior to the blastodermal mandibular anlage (depicted by an open arrow in Figs. 2d, e; 3D; and 6F), at a region that is spatially distinct from the region covered by the early blastoderm expression patterns of hh (Fig. 6E, F) and wg (Fig. 2a, b). A developmentally delayed establishment of the intercalary segment has also been described for Tribolium (Posnien and Bucher 2010; Schaeper et al. 2010), thus reflecting a phylogenetically conserved mode of delayed intercalary segment establishment within at least the holometabolous insects.
Conclusions
Resulting from the functional enhancer dissection assays, the isolation of an intercalary-specific cis-regulatory element of hh supports the concept of a unique mode of establishment of each of the procephalic head segments, as it was initially devised based on the results of mutant analysis (Gallitano-Mendel and Finkelstein 1997). Functional data that add further support to this conclusion are the detection of a large cis-regulatory region of wg which specifically confers establishment of procephalic wg expression only in the antennal segment among the procephalic segments. The above results indicate that distinctive cis-regulatory information and thus segment-specific transcriptional gene networks underlie the metamerization process in the procephalon. This supports the idea that establishment of the procephalic segments reflects the primary segmentation mode (Minelli 2001; Tautz 2004). Moreover, regulatory mechanisms comprising enhancer–promoter-specific interactions and the function of additional temporal cis-regulatory control elements contribute to the specificity of transcriptional regulation governing the segment polarity gene expression in the anterior head. The functional isolation of an intercalary-specific cis-regulatory element of hh can now lead to the identification and verification of direct, trans-acting sequence-specific binding factors and thus elucidate how the patterning information is molecularly transmitted from the head gap-like genes to segment polarity gene expression. Two candidates for such second order regulators acting on this particular ic-CRE are the helix-loop-helix transcription factor Collier (Crozatier et al. 1996, 1999) and the basic-leucine-zipper transcription factor Cap’n’collar B (Mohler et al. 1995; Veraksa et al. 2000). A detailed, molecular, and biochemical analysis on the direct interactions of these second order regulators with the ic-CRE will be published elsewhere (EN and EAW unpublished). Finally, our results support that during the process of metamerization of the anterior head region, the establishment of the intercalary segment — apart from having a morphogenetic independent origin — is also developmentally delayed in comparison to the rest of the procephalic segments. The latter also provides an indication for an evolutionary conserved mode of establishment of the insect intercalary segment (Posnien and Bucher 2010; Schaeper et al. 2010).
Acknowledgments
We would like to thank Dr. Michele Calos, Stanford University for the pTA-attB plasmid; Dr. Johannes Bischof, University of Zurich for providing us with fly lines of the site-specific integration system; and Frank Pogoda for cloning pLSfa_hs43_lacZ_af. This work has been supported by the European Community’s Marie Curie Research Training Network ZOONET under contract MRTN-CT-2004-005624 (to EAW).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Evgenia Ntini, Email: entini@gwdg.de.
Ernst A. Wimmer, Phone: +49-551-3922889, FAX: +49-551-395416, Email: ewimmer@gwdg.de
References
- Abzhanov A, Kaufman TC. Homeotic genes and the arthropod head: expression patterns of the labial, proboscipedia, and Deformed genes in crustaceans and insects. Proc Natl Acad Sci USA. 1999;96:10224–10229. doi: 10.1073/pnas.96.18.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Siepel AC, Kent WJ, Haussler D. Computational screening of conserved genomic DNA in search of functional noncoding elements. Nat Meth. 2005;2:535–545. doi: 10.1038/nmeth0705-535. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher G, Wimmer EA. Beetle a-head. B.I.F. Futura. 2005;20:164–169. [Google Scholar]
- Butler JE, Kadonaga JT. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melenogaster. 2. Berlin: Springer; 1997. [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Cerny AC, Grossmann D, Bucher G, Klingler M. The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Dev Biol. 2008;321:284–294. doi: 10.1016/j.ydbio.2008.05.527. [DOI] [PubMed] [Google Scholar]
- Chang T, Mazotta J, Dumstrei K, Dumitrescu A, Hartenstein V. Dpp and Hh signaling in the Drosophila embryonic eye field. Development. 2001;128:4691–4704. doi: 10.1242/dev.128.23.4691. [DOI] [PubMed] [Google Scholar]
- Chipman AD, Akam M. The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair-rule gene homologues. Dev Biol. 2008;319:160–169. doi: 10.1016/j.ydbio.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. Genetic regulation of engrailed and wingless in Tribolium segmentation and the evolution of pair-rule segmentation. Dev Biol. 2009;325:482–491. doi: 10.1016/j.ydbio.2008.10.037. [DOI] [PubMed] [Google Scholar]
- Choe CP, Miller SC, Brown SJ. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci USA. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill ME, Jones DN, Glaser T, Hefner H, Searles MA, Travers AA. HMG-D is an architecture-specific protein that preferentially binds to DNA containing the dinucleotide TG. EMBO J. 1995;14:1264–1275. doi: 10.1002/j.1460-2075.1995.tb07110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Jürgens G. Mediation of Drosophila head development by gap-like segmentation genes. Nature. 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- Cohen S, Jürgens G. Drosophila headlines. Trends Genet. 1991;7:267–272. doi: 10.1016/0168-9525(91)90327-M. [DOI] [PubMed] [Google Scholar]
- Crémazy F, Berta P, Girard F. Genome-wide analysis of Sox genes in Drosophila melanogaster. Mech Dev. 2001;109:371–375. doi: 10.1016/S0925-4773(01)00529-9. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–718. doi: 10.1016/S0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Head versus trunk patterning in the Drosophila embryo; collier requirement for formation of the intercalary segment. Development. 1999;126:4385–4394. doi: 10.1242/dev.126.19.4385. [DOI] [PubMed] [Google Scholar]
- Dailey L, Basilico C. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J Cell Physiol. 2001;186:315–328. doi: 10.1002/1097-4652(2001)9999:9999<000::AID-JCP1046>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Damen WG. Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. Development. 2002;129:1239–1250. doi: 10.1242/dev.129.5.1239. [DOI] [PubMed] [Google Scholar]
- de Velasco B, Mandal L, Mkrtchyan M, Hartenstein V. Subdivision and developmental fate of the head mesoderm in Drosophila melanogaster. Dev Genes Evol. 2006;216:39–51. doi: 10.1007/s00427-005-0029-4. [DOI] [PubMed] [Google Scholar]
- Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill ME, Crane-Robinson C, Privalov PL. DNA binding and bending by HMG boxes: energetic determinants of specificity. J Mol Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Dray N, Tessmar-Raible K, Le Gouar M, Vibert L, Christodoulou F, Schipany K, Guillou A, Zantke J, Snyman H, Béhague J, Vervoort M, Arendt D, Balavoine G. Hedgehog signaling regulates segment formation in the annelid Platynereis. Science. 2010;329:339–342. doi: 10.1126/science.1188913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallitano-Mendel A, Finkelstein R. Novel segment polarity gene interactions during embryonic head development in Drosophila. Dev Biol. 1997;192:599–613. doi: 10.1006/dbio.1997.8753. [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci USA. 1997;94:12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Cadigan KM, Gehring WJ. Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development. 1994;120:3155–3171. doi: 10.1242/dev.120.11.3155. [DOI] [PubMed] [Google Scholar]
- Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210:630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- Horn C, Jaunich B, Wimmer EA. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev Genes Evol. 2000;210:623–629. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Juven-Gershon T, Marr MT, 2nd, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Kamachi Y. SOX-partner code for cell specification: regulatory target selection and underlying molecular mechanisms. Int J Biochem Cell Biol. 2010;42:391–399. doi: 10.1016/j.biocel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/MCB.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-D. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing D, Nusse R. Expression of wingless in the Drosophila embryo: a conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development. 1998;125:1469–1476. doi: 10.1242/dev.125.8.1469. [DOI] [PubMed] [Google Scholar]
- Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang X, Dong Y, Friedrich M. Tracking down the “head blob”: comparative analysis of wingless expression in the developing insect procephalon reveals progressive reduction of embryonic visual system patterning in higher insects. Arthropod Struct Dev. 2006;35:341–356. doi: 10.1016/j.asd.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- Maderspacher F, Bucher G, Klingler M. Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev Genes Evol. 1998;208:558–568. doi: 10.1007/s004270050215. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-N. [DOI] [PubMed] [Google Scholar]
- McKimmie C, Woerfel G, Russell S. Conserved genomic organisation of group B Sox genes in insects. BMC Genet. 2005;6:26. doi: 10.1186/1471-2156-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A. A three-phase model of arthropod segmentation. Dev Genes Evol. 2001;211:509–521. doi: 10.1007/s004270100180. [DOI] [PubMed] [Google Scholar]
- Mohler J. Spatial regulation of segment polarity gene expression in the anterior terminal region of the Drosophila blastoderm embryo. Mech Dev. 1995;50:151–161. doi: 10.1016/0925-4773(94)00332-H. [DOI] [PubMed] [Google Scholar]
- Mohler J, Mahaffey JW, Deutsch E, Vani K. Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development. 1995;121:237–247. doi: 10.1242/dev.121.1.237. [DOI] [PubMed] [Google Scholar]
- Nambu PA, Nambu JR. The Drosophila fish-hook gene encodes a HMG domain protein essential for segmentation and CNS development. Development. 1996;122:3467–3475. doi: 10.1242/dev.122.11.3467. [DOI] [PubMed] [Google Scholar]
- Ntini E (2009) Regulation of segment polarity genes in the head region of different arthropods. Dissertation, Georg-August-University Göttingen
- Onichtchouk D, Geier F, Polok B, Messerschmidt DM, Mössner R, Wendik B, Song S, Taylor V, Timmer J, Driever W. Zebrafish Pou5f1-dependent transcriptional networks in temporal control of early development. Mol Syst Biol. 2010;6:354. doi: 10.1038/msb.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Developmental evolution: insights from studies of insect segmentation. Science. 1994;266:581–590. doi: 10.1126/science.7939712. [DOI] [PubMed] [Google Scholar]
- Pechmann M, McGregor AP, Schwager EE, Feitosa NM, Damen WG. Dynamic gene expression is required for anterior regionalization in a spider. Proc Natl Acad Sci USA. 2009;106:1468–1472. doi: 10.1073/pnas.0811150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posnien N, Bucher G. Formation of the insect head involves lateral contribution of the intercalary segment, which depends on Tc-labial function. Dev Biol. 2010;338:107–116. doi: 10.1016/j.ydbio.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Rehm EJ, Hannibal RL, Chaw RC, Vargas-Vila MA, Patel NH. In situ hybridization of labeled RNA probes to fixed Parhyale hawaiensis embryos. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5130. [DOI] [PubMed] [Google Scholar]
- Reichert H, Simeone A. Conserved usage of gap and homeotic genes in patterning the CNS. Curr Opin Neurobiol. 1999;9:589–595. doi: 10.1016/S0959-4388(99)00002-1. [DOI] [PubMed] [Google Scholar]
- Remane A. Enstehung der Metamerie der Wirbellosen. Zool Anz Suppl. 1950;14:16–23. [Google Scholar]
- Rogers BT, Kaufman TC. Structure of the insect head in ontogeny and phylogeny: a view from Drosophila. Int Rev Cytol. 1997;174:1–84. doi: 10.1016/S0074-7696(08)62115-4. [DOI] [PubMed] [Google Scholar]
- Russell SR, Sanchez-Soriano N, Wright CR, Ashburner M. The Dichaete gene of Drosophila melanogaster encodes a SOX-domain protein required for embryonic segmentation. Development. 1996;122:3669–3676. doi: 10.1242/dev.122.11.3669. [DOI] [PubMed] [Google Scholar]
- Sánchez-Soriano N, Russell S. Regulatory mutations of the Drosophila Sox gene Dichaete reveal new functions in embryonic brain and hindgut development. Dev Biol. 2000;220:307–321. doi: 10.1006/dbio.2000.9648. [DOI] [PubMed] [Google Scholar]
- Sander K. The evolution of patterning mechanisms: gleanings from insect embryogenesis and spermatogenesis. In: Goodwin BC, Holder N, Wylie CC, editors. Development and evolution. Cambridge: Cambridge University Press; 1983. pp. 137–160. [Google Scholar]
- Schaeper ND, Pechmann M, Damen WG, Prpic NM, Wimmer EA. Evolutionary plasticity of collier function in head development of diverse arthropods. Dev Biol. 2010;344:363–376. doi: 10.1016/j.ydbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Schinko JB, Kreuzer N, Offen N, Posnien N, Wimmer EA, Bucher G. Divergent functions of orthodenticle, empty spiracles and buttonhead in early head patterning of the beetle Tribolium castaneum (Coleoptera) Dev Biol. 2008;317:600–613. doi: 10.1016/j.ydbio.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, Technau GM. Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development. 1992;116:111–125. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, González-Gaitán M, Jäckle H, Technau GM. Number, identity, and sequence of the Drosophila head segments as revealed by neural elements and their deletion patterns in mutants. Proc Natl Acad Sci USA. 1994;91:8363–8367. doi: 10.1073/pnas.91.18.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz G. Evolution of the nauplius stage in malacostracan crustaceans. J Zoolog Syst Evol Res. 2000;38:175–187. doi: 10.1046/j.1439-0469.2000.383151.x. [DOI] [Google Scholar]
- Scholtz G, Edgecombe GD. The evolution of arthropod heads: reconciling morphological, developmental and palaeontological evidence. Dev Genes Evol. 2006;216:395–415. doi: 10.1007/s00427-006-0085-4. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-P. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Segmentation. Dev Cell. 2004;7:301–312. doi: 10.1016/j.devcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Pirotta V. New pCaSpeR P-element vectors. Drosoph Inf Serv. 1992;71:150. [Google Scholar]
- Urbach R, Technau GM. Early steps in building the insect brain: neuroblast formation and segmental patterning in the developing brain of different insect species. Arthropod Struct Dev. 2003;32:103–123. doi: 10.1016/S1467-8039(03)00042-2. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/S0092-8674(00)81925-X. [DOI] [PubMed] [Google Scholar]
- Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap ‘n’ collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- Vincent A, Blankenship JT, Wieschaus E. Integration of the head and trunk segmentation systems controls cephalic furrow formation in Drosophila. Development. 1997;124:3747–3754. doi: 10.1242/dev.124.19.3747. [DOI] [PubMed] [Google Scholar]
- Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X, Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- Wilson M, Koopman P. Matching SOX partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/S0959-437X(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Wimmer EA, Cohen SM, Jäckle H, Desplan C. buttonhead does not contribute to a combinatorial code proposed for Drosophila head development. Development. 1997;124:1509–1517. doi: 10.1242/dev.124.8.1509. [DOI] [PubMed] [Google Scholar]
- Wolff C, Sommer R, Schröder R, Glaser G, Tautz D. Conserved and divergent expression aspects of the Drosophila segmentation gene hunchback in the short germ band embryo of the flour beetle Tribolium. Development. 1995;121:4227–4236. doi: 10.1242/dev.121.12.4227. [DOI] [PubMed] [Google Scholar]


