Abstract
Context
Glioblastomas—uniformly fatal brain tumors—often have both monosomy of chromosome 10 and gains of the epidermal growth factor receptor (EGFR) gene locus on chromosome 7, an association for which the mechanism is poorly understood.
Objectives
To assess whether coselection of EGFR gains on 7p12 and monosomy 10 in glioblastomas promotes tumorigenic epidermal growth factor (EGF) signaling through loss of the annexin A7 (ANXA7) gene on 10q21.1–q21.2 and whether ANXA7 acts as a tumor suppressor gene by regulating EGFR in glioblastomas.
Design, Setting, and Patients
Multidimensional analysis of gene, coding sequence, promoter methylation, messenger RNA (mRNA) transcript, protein data for ANXA7 (and EGFR), and clinical patient data profiles of 543 high-grade gliomas from US medical centers and The Cancer Genome Atlas pilot project (made public 2006–2008; and unpublished, tumors collected 2001–2008). Functional analyses using LN229 and U87 glioblastoma cells.
Main Outcome Measures
Associations among ANXA7 gene dosage, coding sequence, promoter methylation, mRNA transcript, and protein expression. Effect of ANXA7 haploinsufficiency on EGFR signaling and patient survival. Joint effects of loss of ANXA7 and gain of EGFR expression on tumorigenesis.
Results
Heterozygous ANXA7 gene deletion is associated with significant loss of ANXA7 mRNA transcript expression (P=1×10−15; linear regression) and a reduction (mean [SEM]) of 91.5% (2.3%) of ANXA7 protein expression compared with ANXA7 wild-type glioblastomas (P=.004; unpaired t test). ANXA7 loss of function stabilizes the EGFR protein (72%–744% increase in EGFR protein abundance) and augments EGFR transforming signaling in glioblastoma cells. ANXA7 haploinsufficiency doubles tumorigenic potential of glioblastoma cells, and combined ANXA7 knockdown and EGFR overexpression promotes tumorigenicity synergistically. The heterozygous loss of ANXA7 in≈75% of glioblastomas in the The Cancer Genome Atlas plus infrequency of ANXA7 mutation (≈6% of tumors) indicates its role as a haploinsufficiency gene. ANXA7 mRNA transcript expression, dichotomized at the median, associates with patient survival in 191 glioblastomas (log-rank P=.008; hazard ratio [HR], 0.667; 95% confidence interval [CI], 0.493–0.902; 46.9 vs 74.8 deaths/100 person-years for high vs low ANXA7 mRNA expression) and with a separate group of 180 high-grade gliomas (log-rank P=.00003; HR, 0.476; 95% CI, 0.333–0.680; 21.8 vs 50.0 deaths/100 person-years for high vs low ANXA7 mRNA expression). Deletion of the ANXA7 gene associates with poor patient survival in 189 glioblastomas (log-rank P=.042; HR, 0.686; 95% CI, 0.476–0.989; 54.0 vs 80.1 deaths/100 person-years for wild-type ANXA7 vs ANXA7 deletion).
Conclusion
Haploinsufficiency of the tumor suppressor ANXA7 due to monosomy of chromosome 10 provides a clinically relevant mechanism to augment EGFR signaling in glioblastomas beyond that resulting from amplification of the EGFR gene.
Aneuploidy—an abnormal number of chromosomes—is the most frequently identified genomic abnormality in human cancer. Cancer-specific aneuploidies often result in activation of oncogenes or loss of tumor suppressor genes.1 Conserved aneuploidies also shape the genome of glioblastomas,2 which are among the most challenging of human cancers. The most frequent form of aneuploidy in glioblastomas is monosomy of chromosome 10, ie, the presence of only 1 chromosome 10 due to loss of 1 copy. We have shown monosomy of chromosome 10 to be significantly associated with gain or amplification of the epidermal growth factor receptor gene (EGFR [HGNC 3236]) on chromosome 7p12,2 the most common genetic defect in growth factor signaling in these tumors.
This association suggests a fundamental biological role in glioblastoma pathogenesis, yet its molecular basis is poorly understood. Conceivably, the concurrent gain of EGFR and monosomy of chromosome 10 could select for a synergistic mechanism to alter EGFR transforming signaling. Such synergy could increase tumorigenic fitness and thus provide the selective pressure to conserve monosomy of chromosome 10 in glioblastomas. Based on integrative analyses of multidimensional genomic, clinical, and interactome (BOX) data and network modeling, we have pinpointed gene residents on chromosome 10 with putative reciprocal relationships to EGFR.2 A new relationship that emerged from this analysis involves EGFR and the potential tumor suppressor gene annexin A7 (ANXA7 [HGNC 545])3 on 10q21.1–q21.2. ANXA7, a calcium-dependent guanosine triphosphatase that binds phospholipid-containing membranes,4,5 has been implicated as a candidate tumor suppressor gene in breast and prostate cancer,6–10 but its cancer-associated biology remains poorly understood.11
Haploinsufficiency (Box) occurs when a diploid organism (2 copies of each chromosome), such as a human, has only 1 functional copy of a gene that does not produce enough protein to sustain a wild-type phenotype.12 In cancer biology, a tumor suppressor gene is termed haploinsufficient if the mutation or loss of only 1 allele (Box) is sufficient to induce a cellular phenotype that leads to tumorigenesis without inactivation of the wild-type allele.13 Haploinsufficiency of ANXA7 has been shown to be pertinent in cancer cells11; reduced production of ANXA7 as a result of monosomy of chromosome 10 could thus diminish the ability of ANXA7 to function properly in glioblastomas.2
We hypothesize that haploinsufficiency of ANXA7 provides an additional mechanism for tumorigenic increase in EGFR signaling in glioblastomas, and thus offers an explanation for the conservation of monosomy 10 in these tumors. We investigated the mechanism of deregulation of ANXA7 in glioblastomas, its link to EGFR signaling, and its association with patient outcome.
METHODS
Tumor Samples
Fifty-nine snap-frozen glioma specimens and 25 formalin-fixed paraffin-embedded glioma specimens were collected at Stanford University, Palo Alto, California, and Northwestern University, Chicago, Illinois, respectively, under institutional review board–approved guidelines and subjected to standard World Health Organization (WHO) classification.14 Written informed consent was obtained from all patients. Specimens were analyzed by a neuropathologist to confirm the histological diagnosis and the presence of vital tumor tissue without excessive contamination (<10%) by normal brain and tumor necrosis. Genomic DNA was isolated using the DNeasy tissue kit (Qiagen, Germantown, Maryland), DpnII restriction endonuclease-digested (New England BioLabs, Beverly, Massachusetts), and purified using the QIA-quick PCR purification kit (Qiagen). Genomic DNA was also extracted from a postmortem human normal brain tissue. Normal human karyotype DNA was purchased from Promega (Madison, Wisconsin).
Agilent human genome CGH microarray 244A gene dosage data (Agilent Technologies, Santa Clara, California), Affymetrix HT human genome U133A array plate set messenger RNA (mRNA) transcript expression data (Affymetrix, Santa Clara, California), and clinical data for 219, 188, and 207 glioblastomas, respectively, were obtained from the open-access and controlled-access data tiers portal (http://tcga-data.nci.nih.gov/tcga/findArchives.htm) of The Cancer Genome Atlas (TCGA) pilot project (http://cancergenome.nih.gov/index.asp) based on National Human Genome Research Institute approval. At the time of data retrieval from TCGA, there was incomplete overlap in the types of data available for each sample. Alignment of sample identifiers yielded 189 samples with gene dosage and clinical data, 175 samples with mRNA transcript expression and clinical data, 175 samples with gene dosage and mRNA transcript expression data, and 172 samples with all 3 data types.
Raw Agilent human genome CGH microarray 244A data were background corrected using subtraction and then replacement of negative or zero values with half the minimum of the positive corrected values on the array and then locally weighted least squares (LOWESS) normalized using the “limma” package for R statistical software (R Foundation for Statistical Computing [http://www.r-project.org/]). Circular binary segmentation (CBS) from the R package “snapCGH” was used to estimate segmented regions of equal dosage along each chromosome. Gene dosage segments were classified as having chromosomal gain or loss if the absolute value of the predicted dosage was more than 0.75 times the interquartile range of the difference between observed and predicted values for each region. The downloaded mRNA transcript expression data were background corrected and normalized using the robust multigene average algorithm15 as described by TCGA.16 For survival analysis, a unified data set of genome-scale expression data for 191 glioblastomas was retrieved from Lee et al.17 Similarly, 2 publicly available high-grade glioma data set studies from the University of Texas M.D. Anderson Cancer Center18 and the University of California, Los Angeles,19 as well as a third unpublished data set from Harvard University, all profiled using the Affymetrix human genome U133A array platform, were analyzed as a unified data set.
Cell Lines and Cell Culture
Glioblastoma cell lines LN229 and U87 were obtained from the American Type Culture Collection (Manassas, Virginia) and were cultured in a humidified incubator at 37°C and 5% CO2 with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Carlsbad, California)–supplemented Dulbecco's Modified Eagle Medium (DMEM, Mediatech Inc, Herndon, Virginia). Primary brain tumor stem cell cultures were generated from fresh glioblastoma tissue by enzymatic dissociation using collagenase and deoxyribonuclease. Tumor cells were resuspended at a density of 5×104 cells/mL in serum-free DMEM/F12 with N2 and B27 nutrient additives supplemented with epidermal growth factor (EGF, 20 ng/mL), fibroblast growth factor (20 ng/mL) (BD Biosciences, San Jose, California), and leukemia-inhibiting factor (10 ng/mL) (Millipore, Billerica, Massachusetts). Tumor spheres were usually detectable within the first week in vitro. For passaging, spheres were dissociated using Accutase (Millipore) and subcultured as previously detailed. Fetal neural stem cells were obtained and maintained as described.20 Adult human astrocytes were harvested from temporal lobe tissue obtained during anterior temporal lobectomy with amygdalo-hippocampectomy and grown in DMEM/F12 supplemented with 10% fetal bovine serum.
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization analysis for chromosomal and extrachromosomal EGFR gene dosage was performed according to previously described methods21 using a probe targeting the EGFR locus and a probe specific for the centromeric region of chromosome 7 (Spectrum Orange dye and Spectrum Green dye, respectively, Abbott Molecular, Abbott Park, Illinois). Nuclei were counterstained with Hoechst 33342. For image acquisition and analysis, samples were visualized using an Axiovert 200 standard fluorescence microscope (Zeiss, Thornwood, New York) with a 120-W short-arc lamp, excitation and emission filters appropriate for Spectrum Orange (EGFR locus–specific signals), Spectrum Green (centromere 7-specific signals), Hoechst 33342 (nuclear) staining, and X40/X63 objectives (Pan-Neofluar, Zeiss). Images were captured using an Axiocam HRc digital microscope camera (Zeiss), and the accompanying Axiovision 4.5 software (Zeiss).
Protein Extraction and Immunoblotting
Cell lysates from wild-type and transfected LN229 and U87 cells were prepared using the Cell Lysis Buffer (Cell Signaling Technology, Danvers, Massachusetts) containing protease and phosphatase inhibitors (Sigma-Aldrich, St Louis, Missouri), sonicated and total protein was quantitated using Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, Illinois). Blots were exposed to anti-EGFR (Assay Designs, Ann Arbor, Michigan), anti-ANXA7 (Santa Cruz Biotechnology, Santa Cruz, California), antiphospho-ERK1/2 (Thr202/Tyr204; Assay Designs), and anti-α-tubulin (Sigma Aldrich) monoclonal antibodies recognized by an HRP-conjugated horse anti-mouse secondary antibody (Cell Signaling); and antiphospho-EGFR (Tyr1086; Invitrogen), anti-ERK1/2 (Abcam, Cambridge, Massachusetts), and antiphospho-AKT1/2/3 (Ser473; Santa Cruz, California) polyclonal antibodies recognized by an HRP-conjugated goat anti-rabbit secondary antibody (Cell Signaling). Enhanced chemiluminescent reagent (Bio-Rad, Hercules, California) was added to the membranes according to the manufacturer's protocol and visualized by autoradiography. Bands were quantified using ImageJ software version 1.42 (National Institutes of Health, Bethesda, Maryland [http://rsb.info.nih.gov/ij/]) and normalized to α-tubulin loading control.
In Situ Proximity Ligation Assay
We measured ANXA7 protein in tumor samples using a highly sensitive proximity ligation assay.22,23 For sample pretreatment, 4-μm-thick formalin-fixed paraffin-embedded tumor sections or a 4-μm-thick tissue microarray were deparaffinized then epitope-retrieved in Citrate Buffer pH6.0 (DBS, Pleasanton, California) under pressure boiling for 5 minutes. After an additional 40 minutes and cooling to room temperature, slides were permeabilized with a 0.1% Triton X-100 solution and rinsed in phosphate-buffered saline. The Duolink procedure (Duolink PLA Kit; Olink, Uppsala, Sweden) was followed as directed by the manufacturer. Slides were incubated with mouse anti-Annexin A7 primary antibody (2 ng/μL; Santa Cruz Biotechnology) at 4°C. Number 1 glass coverslips (VWR International, West Chester, Pennsylvania) were mounted on samples with Vectashield Mounting Medium (Vector Laboratories, Burlingame, California).
For image acquisition and analysis, samples were visualized using an Axiovert 200 standard fluorescence microscope (Zeiss) with a 120-W short-arc lamp, excitation and emission filters appropriate for Texas Red (proximity ligation assay signals) and Hoechst 33342 (nuclear) staining, and an X40 objective (Pan-Neofluar; Zeiss). Images were obtained using an Axiocam HRc and Axiovision 4.5 software. For quantification of ANXA7 protein, images were converted to 8-bit grayscale tag image file format (TIF) images and analyzed using BlobFinder software V3.0 (Uppsala University, Uppsala, Sweden [www.cb.uu.se/~amin/BlobFinder/]) to provide the total number of proximity ligation assay (single protein) signals and the total number of nuclei using average signal count analysis (after optimization and standardization of nucleus size, blob size, blob intensity, and cytoplasm radius). Images were obtained using an LSM 510 laser scanning confocal microscope (Zeiss).
Bisulfite Treatment and Polymerase Chain Reaction
Bisulfite modification of genomic DNA was performed using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, California) according to manufacturer's instructions (0.5 μg of DNA was bisulfite-treated, eluted with 20 μL elution solution, and stored at −20°C until ready for use). Nested polymerase chain reaction (PCR) reactions were adopted to amplify the ANXA7 gene promoter region. Two runs of PCR reactions were carried out using the Hotstart Taq polymerase kit (Qiagen) in 25 μL total volume and with 50 pM of forward primer and reverse primer. In the first PCR reaction, 50 ng of the bisulfite-converted DNA in 1 μL was used as a template. After 3 minutes of initial denaturation at 95°C, the cycling conditions of 25 cycles consisted of denaturation at 95°C for 15 seconds, annealing at 50°C for 15 seconds, and elongation at 72°C for 15 seconds. One microliter of PCR product from the first run was used as the template for the second PCR reaction, which after initial denaturation at 95°C for 3 minutes comprised 45 cycles with denaturation at 95°C for 15 seconds, annealing at 47°C for 15 seconds, and elongation at 72°C for 15 seconds. The primers used in the first PCR run were 5–GAG GTG GGA AAG GGAGG-3′ and 5′-AAA AAT CTT TTT CCC TCA AAT AAC-3′; those of the second PCR run were 5′-AAA CTA AAA AAC AAC CC-3′ and biotinylated primer 5′-/5Biosg/TTT TTT ATT TGA TTT TAG TAG TAG-3′. The PCR products were stored at 4°C until ready for pyrosequencing.
Pyrosequencing
Pyrosequencing, a method of DNA sequencing based on the sequencing by synthesis principle24 that can accurately quantify methylation patterns at individual nucleotide positions, was performed as previously described25 using the PyroMark MD pyrosequencing system (Biotage, Charlottesville, Virginia). The final PCR product was purified using streptavidin-Sepharose HP beads (GE Healthcare, Uppsala, Sweden) and processed to yield single-stranded DNA. The single-stranded DNA was prepared for pyrosequencing using the PyroMark vacuum prep tool (Biotage). The PCR product was bound onto Sepharose beads. Beads containing the immobilized PCR product were washed, denatured using a 0.2M NaOH solution, washed again, and neutralized. Pyrosequencing primer at a concentration of 0.3 μM was annealed to the purified single-stranded PCR product at 28°C. The primer sequence to determine the methylation level of 9 CpG sites within the ANXA7 promoter located close to the transcriptional start site was 5′-ACT AAA AAA CAA CCC-3′. Methylation quantification was performed using the manufacturer-provided software (Biotage) with default settings.
Retroviral Infection and EGF Stimulation
Retroviral production was performed in tsA54 packaging cells as previously described.26,27 Human short hairpin (sh)ANXA7 knockdown retroviral construct in a pSM2 vector (Open Biosystems, Huntsville, Alabama) targeting the 1073–1093 mRNA sequence of ANXA7 was cotransfected with pIK packaging vector. Human empty retroviral vector pBabe (EV) was used as a retroviral control. For stable ANXA7 knockdown, U87 cells were retrovirally infected for 2 days, twice a day for 4 hours, in the presence of 5 to 10 μg/mL Polybrene (Sigma-Alrich), followed by 3 days of 1 μg/mL Puromycin selection (Calbiochem, San Diego, California). Transduced cells were stably maintained in 0.5 μg/mL Puromycin selection. For transient transfection, LN229 and U87 cells were transfected with shANXA7 pSM2 and N-terminal FLAG-tagged EGFR pcDNA3.1 plasmids using Lipofectamine 2000 (Invitrogen). Transfected cell lines were selected after 2 to 3 passages over 15 days in 250 μg/mL G418 (Calbiochem) and 1 μg/mL Puromycin. For epidermal growth factor (EGF) stimulation experiments following 24 hours serum starvation, U87-ANXA7 knockdown and U87-EV cells were stimulated with 50 ng/mL recombinant human EGF (Sigma-Alrich) and assayed at various time points (0-, 30-, 60-, and 90-minute) for total and phosphorylated EGFR, extracellular signal-regulated kinase 1/2 (ERK1/2), and V-akt murine thymoma viral oncogene homolog 1/2/3 (phosphorylated AKT1/2/3) abundance by immunoblotting.
Real-Time Reverse Transcription PCR
Quantitative real-time reverse transcription PCR (qRT-PCR) reactions were performed with the ABI 7900HT Fast Real-Time PCR System and Sequence Detection Software V2.3 (Applied Biosystems, Foster City, California) using prevalidated TaqMan Gene Expression Assays (Applied Biosystems) designed to amplify the EGFR mRNA transcript and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping mRNA transcript. Total RNA (5 μg) from U87-EV, LN229-EV, U87-shANXA7, and LN229-shANXA7 cells was extracted using the RNeasy Mini Kit (Qiagen) and reverse transcribed using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). Thermocycling for each PCR reaction was carried out in a final volume of 20 μL containing 100 ng of complementary DNA, 1 TaqMan MGB probe (6-FAM dye-labeled), 250 nM final concentration, and 1× TaqMan Universal PCR Master Mix. After 10 minutes of initial denaturation at 95°C, the cycling conditions of 40 cycles consisted of denaturation at 95°C for 15 seconds followed by annealing and extension at 60°C for 1 minute. All reactions were performed in triplicate. Relative quantitation studies were carried out in RQ Manager Software version 1.2 (Applied Biosystems) using the ΔΔ threshold cycle (CT) method.
Soft-Agar Assay
Retrovirally transfected/transduced LN229 and U87 cells were trypsinized, counted, and suspended in 0.2% agar in 20% fetal bovine serum DMEM, and plated in duplicate on 60-mm dishes containing 0.5% agarose at 1×105 cells per plate. DMEM medium was changed every alternate day. Colonies were observed and counted at 3 different sites at various time points (day 15 to 35), stained for viability with 3 mg/mL 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye (Calbiochem) for 4 hours, and visualized with a standard light microscope. Colony formation indicates that individual cells develop into cell clones that are identified as single colonies. Colony formation or colony-forming activity is a phenotypically recognizable characteristic of cell transformation and a measure of malignant tumor cell behavior.
Mutational Analysis
Sequencing data and inferred mutations generated at the Broad Institute, Cambridge, Massachusetts were obtained from TCGA. At the time of this report, mutations were reported for 149 unique tumor samples. In this project, the Sanger/di-deoxy method was used to sequence whole genome–amplified-genomic DNA from tumor and paired normal samples. After correcting for the background nucleotide-specific mutation rate and sequence coverage of each gene, mutations were called (ie, identified as missense mutations, frame-shift mutations, etc) and then verified by comparison with DNA sequence derived from normal tissue from the same patient using a second genotyping platform. Furthermore, a recent genome-wide mutational study of glioblastoma28 was queried for reported ANXA7 mutations, which has sequenced the protein coding exons of 23 219 mRNA transcripts representing 20 661 genes in 22 glioblastoma samples using next-generation sequencing technology.
Statistics and Survival Analysis
All statistical calculations were performed using R version 2.8.1 and packages from the Bioconductor release 2.3 (Bioconductor, http://www.bioconductor.org/). Unless otherwise stated, statistical significance was established according to 2-sided P values of less than .05 for all statistical tests.
Linear regression analysis was used to assess the relationship between gene dosage and mRNA transcript expression for ANXA7 in the TCGA set and for the promoter methylation/mRNA expression and gene dosage/mRNA expression relationships in the Stanford set, using mRNA transcript expression as the outcome and gene dosage or promoter methylation as the predictor. Scatterplots and LOWESS smooths were used to confirm the suitability of linear regression analyses, and statistical significance of these relationships was assessed according to the P value for the estimated slope of the regression line. The unpaired t test and Wilcoxon rank-sum test were used as appropriate.
Survival curves between groups were estimated by the Kaplan-Meier product-limit method and survival distributions between groups were compared using the Mantel-Cox log-rank test. Univariate and multivariate Cox proportional hazards regression analyses were performed with overall duration of survival as the dependent variable and gene dosage or mRNA transcript expression as the primary predictor of interest. The assumption of proportional hazards was tested using interactions of the predictor variables with time. The circular binary segmentation and robust multigene average preprocessing algorithms yield continuous gene dosage and expression estimates on a log2 scale of red (Cy5 dye) and green (Cy3 dye) fluorescence intensities (specifically ratios of Cy5 to Cy3 dyes or log2R/G), thereby rendering the interpretation of the hazard ratios (HRs) in the Cox models with continuous versions of these predictors somewhat difficult. For easier HR interpretation, we dichotomized mRNA transcript expression at the median and used circular binary segmentation–derived gene dosage loss/wild-type information as binary predictors in the Cox models with overall survival as the outcome.
RESULTS
Patients
Fifty-nine patients treated at Stanford University between April 5, 2001, and April 19, 2004, constituted the initial molecular discovery set. Diagnoses included 36 glioblastomas, 8 astrocytic tumors (WHO grades I–III), 8 oligodendrocytic tumors (WHO grades II and III), and 7 anaplastic oligoastrocytomas (WHO classification14). Twenty-five high-grade glioma patients (21 glioblastomas, 2 anaplastic astrocytomas, and 2 anaplastic oligoastrocytomas) treated at Northwestern University between June 2004 and October 2008 constituted a molecular validation set. There were 219 glioblastoma samples collected between July 26, 1989, and November 23, 2007, and profiled as part of the TCGA pilot project, which constituted an additional molecular validation and a clinical validation set. Corresponding clinical data were available for 207 of the 219 patients (77 females, 130 males), of which 192 were dead and 15 were alive at last follow-up. Mean patient age was 55.8 (SD, 15.1) years. Median duration of follow-up was 50.6 (range, 1.1–503.4) weeks.
A unified collection of 191 glioblastoma patients (74 females, 117 males; made public October 10, 2008) from multiple institutions,17 constituted a second clinical validation set. A total of 163 patients had primary tumors and 28 had recurrent tumors. Mean patient age was 53.8 (SD, 13.6) years; 176 patients were dead and 15 were alive at last follow-up. Median follow-up was 55.6 (range, 1.0–479.0) weeks.
A second unified collection of 180 high-grade glioma patients (85 females, 95 males) from the University of Texas M.D. Anderson Cancer Center18 (made public March 15, 2006), University of California, Los Angeles19 (made public March 8, 2006), and Harvard University (unpublished) constituted a third clinical validation set. There were 129 patients with glioblastomas, 36 with anaplastic astrocytomas, 9 with anaplastic oligodendrogliomas, and 6 with anaplastic oligoastrocytomas. Patient age was 46.5 (SD, 14.6) years. At last follow-up, 128 patients were dead and 58 were alive. Median follow-up was 76.5 (range, 0–492) weeks.
EGFR and ANXA7
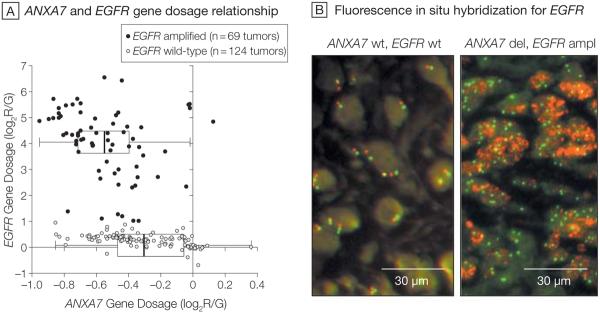
Our characterization of a nonrandom genetic landscape in human gliomas highlighted a significant association between chromosomal alterations containing the EGFR and ANXA7 genes.2 A specific comparison of gene dosage data from TCGA for these 2 genes confirmed a significantly decreased ANXA7 gene dosage for EGFR amplified (n=69; median ANXA7 log2R/G=−0.550) vs EGFR–wild-type tumors (n=124; median ANXA7 log2 R/G=−0.305) (P=1×10−9; Wilcoxon rank-sum test; Figure 1). These analyses included 193 samples for which all EGFR probes on the Agilent aCGH array yielded consistent amplification or wild-type calls. Additional probe-specific analyses using all 219 samples demonstrated similar results.
Figure 1. Relationship Between the ANXA7 and EGFR Genes (193 Tumors).
A, Gene dosage relationship for annexin A7 (ANXA7) and epidermal growth factor receptor (EGFR) in glioblastomas from The Cancer Genome Atlas (TCGA) pilot project. Analysis included 193 of 219 TCGA samples for which all EGFR probes on the Agilent aCGH array yielded consistent amplification or wild-type (wt) calls. Scatterplot shows the relationship between EGFR and ANXA7 gene dosage along the Cartesian coordinates for each of the 193 tumors. Gene dosage values indicate the log2 ratio of red-to-green fluorescence dye intensity (log2R/G) estimated by the circular binary segmentation algorithm. Negative gene dosage indicates lower gene dosage compared with that observed for the majority of the samples, ie, lower gene dosage compared with a normal diploid state. Boxplots depict the first quartile minus 1.5 × interquartile range (IQR) (left whiskers) and the third quartile plus 1.5 × IQR (right whiskers), IQR (box), and median (vertical line); 1 observation outside the whiskers indicates an outlier. Wilcoxon rank-sum testing for the difference in ANXA7 gene dosage in EGFR-amplified (ampl) vs wild-type tumors discloses a significantly diminished ANXA7 gene dosage in the EGFR-amplified tumors (P=.000000001). B, Representative fluorescence in situ hybridization analysis of EGFR gene dosage in 1 ANXA7–wild-type and 1 ANXA7-deleted (del) glioblastoma, from the Stanford set of tumors, on paraffin. Red dots (Spectrum Orange dye) are specific for the EGFR gene locus probe and green dots (Spectrum Green dye) arise from a probe specific for the centromeric region of chromosome 7. The ANXA7–wild-type tumor is nonamplified for EGFR gene dosage with most cells showing normal diploid levels of 2 red and 2 green signals per cell, while the ANXA7-deleted tumor is amplified for EGFR gene dosage as the cells show a robust increase in the number of EGFR red dots. Additionally, the ANXA7-deleted tumor is polyploidic for chromosome 7 as there are more than 2 green dots (centromere of chromosome 7) in most cells (original magnification ×40).
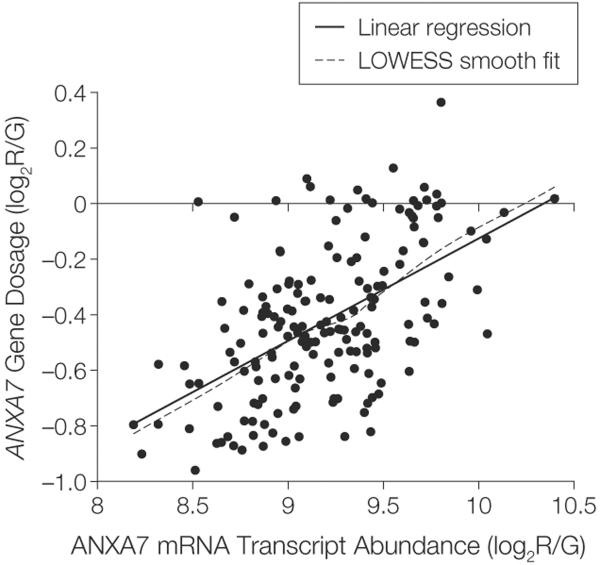
ANXA7 Gene–mRNA Transcript and Gene-Protein Relationships
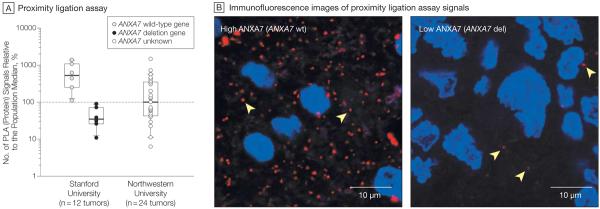
Previous analyses of TCGA pilot project data2 also identified a significant gene dosage-to-expression relationship for ANXA7. Linear regression of ANXA7 mRNA transcript expression on ANXA7 gene dosage in TCGA confirms a significant gene dosage effect on transcription for ANXA7 (P=1×10−15; 174 samples with 1 outlier excluded; Figure 2). We further compared ANXA7 protein abundance in glioblastomas with heterozygous ANXA7 loss (of 1 allele) to that in glioblastomas with wild-type ANXA7 status in a panel of 12 tumors (6 vs 6) from Stanford University. Proximity ligation assay analysis disclosed significantly less ANXA7 protein abundance in tumors with monoallelic ANXA7 loss than in wild-type tumors (P=.004; unpaired t test; Figure 3A). Although none of the ANXA7-deleted tumors had a complete loss of ANXA7 protein expression (indicating a complete gene loss), they showed on average (SEM) a 91.5% (2.3%) reduction of expression compared with the median expression of wild-type tumors (Figure 3B). We extended the ANXA7 proximity ligation assay analysis to a panel of 25 high-grade gliomas from Northwestern University. We were able to analyze ANXA7 protein expression in all but 1 sample (n = 24) that was excluded from the analysis due to red blood cells masking the tumor burden. These 24 tumors showed a spread (range percentage of population median, 6.3–1459.2) of ANXA7 protein expression similar to that of the Stanford set (range, 10.9–1372.8), with a portion of tumors showing very low expression (Figure 3A).
Figure 2. ANXA7 Gene–mRNA Transcript Expression Relationship (175 Tumors).
Relationship between the annexin A7 (ANXA7) gene and ANXA7 mRNA transcript in 175 glioblastomas from The Cancer Genome Atlas with combined availability of gene dosage and expression data for ANXA7. Gene dosage and mRNA expression values are expressed as ratios of red-to-green fluorescence dye intensity (log2R/G) estimated by circular binary segmentation and robust multigene average preprocessing algorithms, respectively. Linear regression of gene expression on gene dosage confirms a significant gene dosage effect on transcription for ANXA7 in 174 tumors (1 outlier excluded) (P=1×10−15). Locally weighted least squares (LOWESS) smooth fit confirmed the appropriateness of a linear regression analysis.
Figure 3. ANXA7 Gene-Protein Relationship.
A, Annexin A7 (ANXA7) protein abundance by proximity ligation assay on paraffin as percentage of protein abundance relative to the population median (horizontal dotted line). The 2 left boxplots compare the ANXA7 protein profiles of 12 glioblastomas from Stanford University (6 ANXA7–wild-type vs 6 ANXA7-deleted tumors); right boxplot reports the ANXA7 protein profiles of 24 high-grade gliomas with unknown ANXA7 gene status from Northwestern University. Boxplots depict the first quartile minus 1.5 × interquartile range (IQR) (lower whiskers) and the third quartile plus 1.5 × IQR (upper whiskers), IQR (box), and median (horizontal line). B, Images of proximity ligation assay demonstrate the range of ANXA7 protein expression in a set of glioblastomas from Stanford University. Left, tumor with highest ANXA7 abundance has ANXA7–wild-type (wt) status; right, tumor with lowest ANXA7 abundance is ANXA7-deleted (del). Proximity ligation assay signals captured by Texas Red and nuclear counterstaining by Hoechst 33342. Arrowheads indicate single ANXA7 proteins (original magnification ×200).
Epigenetic Regulation of ANXA7
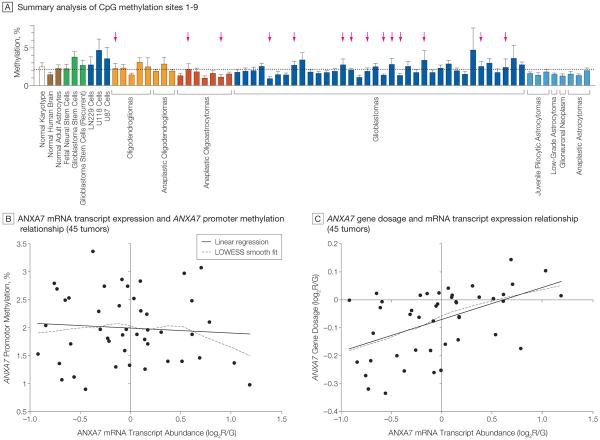
Epigenetic mechanisms—especially differential gene promoter methylation—could also affect the expression of ANXA7 in gliomas. Analysis of the methylation status of 9 CpG sites in the ANXA7 promoter of 59 human glioma samples from patients treated at Stanford University revealed no substantial difference in ANXA7 promoter methylation patterns between various glioma subtypes and both glioma tissue and reference tissues, including normal human karyotype, normal human brain, normal adult astrocytes, and fetal neural and glioblastoma-derived stem cells (Figure 4 A and eFigure available at http://www.jama.com). Linear regression of ANXA7 mRNA transcript expression on ANXA7 promoter methylation (average profile of 9 CpG sites) in 45 Stanford University tumors with combined expression and methylation profiles revealed no significant association (P = .627; Figure 4B). By contrast, linear regression of ANXA7 mRNA transcript expression on ANXA7 gene dosage in the same tumors disclosed a significant gene dosage effect on transcription for ANXA7 (P=.0005; Figure 4C). We found no difference in ANXA7 promoter methylation between ANXA7-deleted tumors (2.07 [mean % SD, 0.81]) vs nondeleted tumors (1.98 [mean % SD, 0.58]); P = .707 by un-paired t test.
Figure 4. ANXA7 Promoter Methylation Analysis and ANXA7 Promoter-Gene–mRNA Transcript Relationship.
A, Pyrosequencing was used to assess the methylation percentage of 9 CpG sites close to the transcriptional start site within the annexin A7 (ANXA7) promoter in 59 human gliomas from Stanford University and various control cells. Bars indicate the overall promoter methylation profile per sample by averaging the methylation profiles of all 9 CpG sites; error bars indicate SD. Horizontal dotted line denotes the average methylation percentage across all samples. Tumors showing deletion of the ANXA7 gene indicated by arrows. Bar colors denote histological subtypes of gliomas. Dark blue coloration of bars for LN229, U118, and U87 cells indicates their origin from glioblastoma tumors. Green bars denote cells with stem cell–like phenotype; brown bars, normal brain tissue and normal brain cells. B, Linear regression of ANXA7 mRNA expression on ANXA7 promoter methylation in 45 of the 59 Stanford University tumors profiled in panel A; availability of corresponding ANXA7 gene expression data reveals no significant relationship (P=.627). The mRNA transcript expression values indicate the log2 ratio of red-to-green fluorescence dye intensity (log2R/G) and are normalized to the median ANXA7 gene expression of all tumors. Locally weighted least squares (LOWESS) smooth fit confirmed the appropriateness of a linear regression analysis. C, Similar linear regression of mRNA transcript expression on gene dosage in the same 45 tumors confirms a significant gene–mRNA transcript relationship for ANXA7 (P=.0005). Gene dosage values are expressed as log2R/G ratios estimated by the circular binary segmentation algorithm. Negative gene dosage indicates lower gene dosage compared with that observed for the majority of the samples, ie, lower gene dosage compared with a normal diploid state.
ANXA7 Loss Augments EGFR and EGFR Signaling
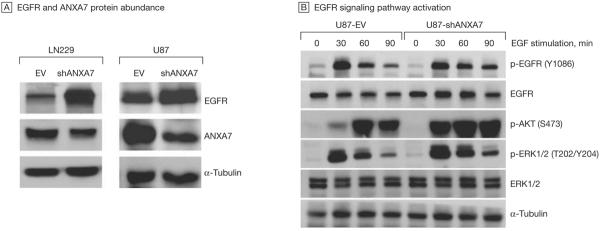
We mimicked a heterozygous gene loss by silencing the ANXA7 gene in LN229 and U87 glioblastoma cells via retroviral transfection of shRNAs targeting ANXA7 mRNA. We found that gene knockdown (Box) of ANXA7 markedly increased EGFR protein abundance (Figure 5A). We found mean (SEM) 744% (34.0%) and 72% (9.3%) increases in total EGFR protein in ANXA7 knockdown LN229 and U87 cells, respectively, compared with corresponding control cells transfected with empty vector only. Furthermore, the knockdown of ANXA7 was associated with increased EGFR activation, resulting in activation of the RAS– mitogen-activated protein kinase and the phosphatidylinositol 3-kinase (PI3K)–AKT pathways, as evidenced by sustained phosphorylation of EGFR at its autophosphorylation site (Tyr1086) and augmented ERK1/2 (at Thr202/Tyr204) and AKT (at Ser473) phosphorylation (Figure 5B).
Figure 5. ANXA7 Knockdown and EGFR Protein Abundance and EGFR Signaling.
A, Knockdown of annexin A7 (ANXA7)—mimicking a hemizygous gene loss and thus haploinsufficiency—via ANXA7-targeting short hairpin RNA (shRNA) (shANXA7) using transient plasmid-based transfection (LN229) or stable, retroviral transduction (U87) increases epidermal growth factor receptor (EGFR) protein abundance compared with cells transfected with empty backbone control vector (EV) only, as assessed by immunoblotting. The α-tubulin protein was used as a loading control. Displayed blots are representative of multiple experiments. B, Total and phosphospecific EGFR, AKT, and ERK1/2 protein abundance in U87-shANXA7 vs U87-EV cells based on immunoblotting. Sustained EGFR autophosphorylation at Tyr 1086 (Y1086) and increased and sustained activating phosphorylation of AKT at Ser473 (S473) and ERK1/2 at Thr202/Tyr204 (T202/Y204) in shANXA7 compared with EV cells.
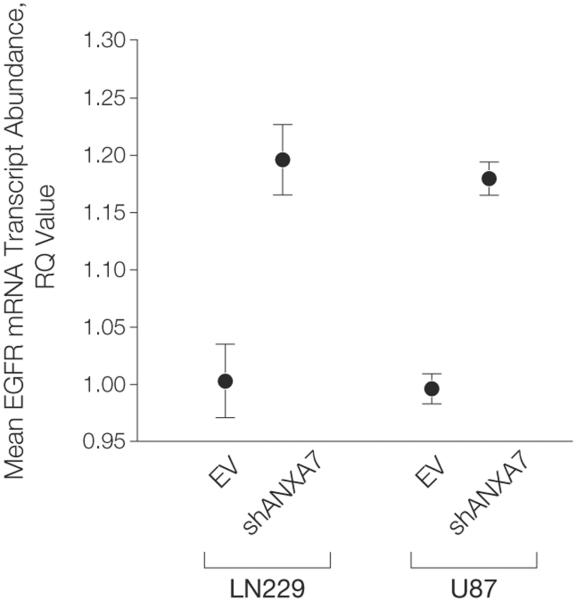
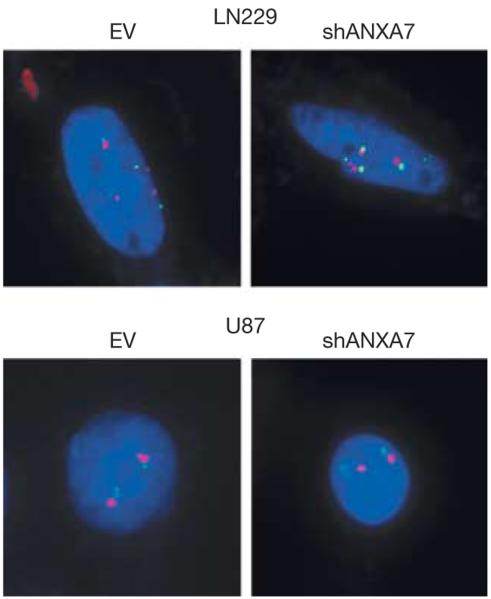
We also studied the effect of ANXA7 knockdown on EGFR mRNA transcript expression in LN229 and U87 cells. We found that ANXA7 knockdown reproducibly resulted in a moderate (mean [SEM], 19.2% [3.1%] and 18.4% [1.5%] for LN229 and U87 cells, respectively) although significant (LN229 and U87, P = .0015 and P=.0002, respectively; unpaired t test) increase in EGFR mRNA transcript expression in both cell lines (Figure 6). This finding is consistent with previous reports suggesting a positive feed-forward circuit of EGFR self-promoted expression in which EGFR activation induces a signaling cascade that increases the expression and stability of its own mRNA transcript.29–31 We further studied the effect of ANXA7 knockdown on the prevalence of EGFR gene dosage located on small fragments of extrachromosomal DNA—so-called double minute chromosomes (Box)—described for EGFR in glioblastomas.32 We found no evidence of increased EGFR gene dosage at its normal chromosomal 7p12 locus or extrachromosomal EGFR gene dosage in ANXA7 knockdown cells: LN229-EV as well as LN229-shANXA7 were tetraploid (4 copies) both for EGFR and chromosome 7, and U87-EV as well as U87-shANXA7 were diploid both for EGFR and chromosome 7 (Figure 7).
Figure 6. ANXA7 Knockdown and EGFR mRNA Transcript Expression.
EGFR (epidermal growth factor receptor) mRNA transcript expression based on quantitative real-time polymerase chain reaction in LN229-EV vs LN229-shANXA7 cells (P=.0015) and in U87-EV vs U87-shANXA7 cells (P=.0002). ANXA7 knockdown results in a moderate though significant increase in EGFR mRNA transcript expression in both cell lines. Error bars denote standard error of the mean. P values according to unpaired t test.
Figure 7. ANXA7 Knockdown and EGFR Gene Dosage.
Fluorescent in situ hybridization analysis detecting the EGFR (epidermal growth factor receptor) gene (red dots, Spectrum Orange dye) and the centromere of chromosome 7 (green dots, Spectrum Green dye) in LN229-EV vs LN229-shANXA7 cells and in U87-EV vs U87-shANXA7 cells. Images show representative gene dosage profiles in single cells. LN229 is tetraploid both for EGFR gene dosage and chromosome 7 with 4 red/green pairs in the nucleus (nuclear counterstaining using Hoechst 33342 stain [blue]). U87 is diploid both for EGFR gene dosage and chromosome 7 with 2 red/green pairs. Both cell lines show no change in EGFR gene dosage on ANXA7 knockdown. The equal number of EGFR-specific and centromeric signals indicates the absence of extrachromosomal EGFR gene dosage in double minute chromosomes, small fragments of extrachromosomal DNA, and potential cytogenetic equivalents of EGFR amplification that contain no centromere (original magnification ×100). ANXA7 indicates annexin A7.
ANXA7 Haploinsufficiency Promotes Glioblastoma Cell Tumorigenicity
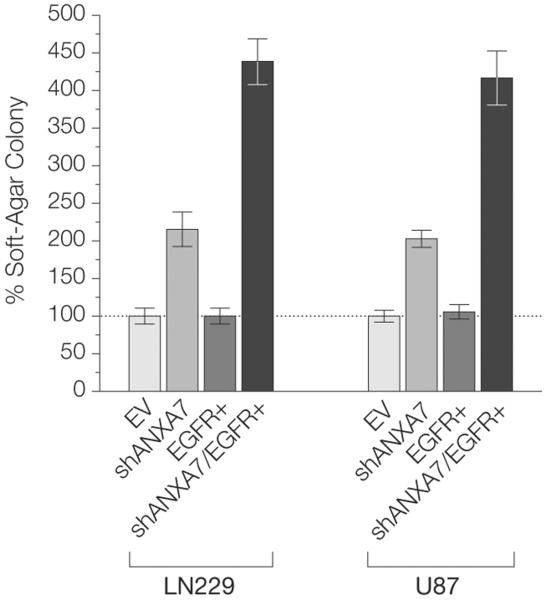
Because of its profound effect on EGFR signaling, we assessed whether loss of ANXA7 function might affect the biological behavior of glioblastoma cells. We found that knockdown of ANXA7 in U87 glioblastoma cells via transfection of ANXA7-targeting shRNA mimicked the loss of 1 allele (50% ANXA7 protein abundance compared with control cells) and thus an ANXA7 haplo-insufficient state (Figure 5A), and resulted in a mean (SEM) 49% (9.8%) increase in colony-forming activity in soft agar (P=.033; unpaired t test) relative to that in cells transfected with control vector only. Pronounced knockdown of ANXA7 (protein levels 20%–25% of control cells) in LN229 and U87 cells led to an even more prominent effect on colony-forming activity compared with that in LN229 and U87 control cells (LN229, P = .0009; U87, P=.0003; unpaired t test), with mean (SEM) increases in colony-forming activity of 215% (20%) and 203% (23%), respectively (Figure 8).
Figure 8. ANXA7 Knockdown and Colony-Forming Activity.
Significantly increased colony-forming activity of shANXA7 cells compared with EV cells (EV is reference) (LN229-shANXA7 vs LN229-EV: P=.0009; U87-shANXA7 vs U87-EV: P = .0003) and of shANXA7/EGFR+ cells compared with shANXA7 cells (LN229-shANXA7/EGFR+ vs LN229-shANXA7: P = .0002; U87-shANXA7/EGFR+ vs U87-shANXA7: P=.001). Error bars denote standard error of the mean colony number of multiple experiments (colony-forming activity also assessed in EV cells). P values according to unpaired t test. ANXA7 indicates annexin A7; EGFR, epidermal growth factor receptor.
Tumorigenic Synergism Between ANXA7 and EGFR
To assess whether there is a cooperative effect on gliomagenesis of combined loss of ANXA7 and gain of EGFR, we compared the tumorigenic potential of glioblastoma cells engineered to express empty control vector, ANXA7 mRNA–targeting shRNA, EGFR, or ANXA7 mRNA–targeting shRNA and EGFR combined in LN229 and U87 cells. Isolated knockdown of ANXA7 again substantially increased the colony-forming activity of glioblastoma cells. EGFR overexpression alone resulted in increased colony sizes but had only a moderate effect on colony numbers. Combined ANXA7 silencing and EGFR overexpression reproducibly amplified the tumor-promoting effect of isolated ANXA7 knockdown in LN229 and U87 cells (P = .0002 and P=.001, respectively; unpaired t test), resulting in mean (SEM) 204% (12%) and 206% (18%) increases in colony-forming activity (Figure 8). These data suggest a tumorigenic synergism between ANXA7 loss and EGFR amplification.
ANXA7 Is a Putative Haploinsufficiency Gene in Glioblastomas
We analyzed the sequencing data for ANXA7 generated by phase I high-throughput sequencing in TCGA to test whether ANXA7 might be a haploin-sufficiency gene in glioblastomas. Out of 15 067 identified mutations across 410 genes in 149 unique TCGA glioblastoma samples, only 11 matched to ANXA7 involving 9 samples (≈6% of all analyzed samples), 4 of which were validated using a second technology. Mutations included 7 missense mutations and 4 frame-shift deletions. A correlation of these mutations with ANXA7 gene dosage status reveals that all tumors (except 1 without dosage data in TCGA) also had ANXA7 deletions. We further queried a recent genome-wide mutational study of 22 glioblastomas, which has sequenced the protein-coding exons of 23 219 mRNA transcripts representing 20 661 genes including exons 1 through 7 and 9 through 12 of ANXA7.28 This study has revealed no somatic mutations in ANXA7 in glioblastomas. These data indicate that small-scale mutations of ANXA7 are rarely present in glioblastomas. Since the frequent (≈75%) deletion of the ANXA7 locus in glioblastomas usually involves the loss of only 1 allele, ANXA7 may indeed function as a haploinsufficiency gene in these tumors.
ANXA7 mRNA Transcript Expression Associated With Malignant Glioma Outcome
To evaluate the effect of ANXA7 haplo-insufficiency on patient outcome, we assessed the association of ANXA7 mRNA transcript expression with the duration of overall survival. In a cohort of 191 glioblastomas,17 ANXA7 mRNA transcript expression was associated with duration of overall survival (HR, 0.74; 95% confidence interval [CI], 0.58–0.95; P=.019; Cox proportional hazards regression), such that loss of expression of ANXA7 denoted a proportionally unfavorable prognosis. A multivariate Cox model incorporating the established prognostic covariates of patient age, disease status (primary vs recurrent), and O6-methylguanine-DNA methyltransferase—currently the most established molecular marker predicting response to standard-of-care treatment in glioblastomas33—sustained an independent association between duration of patient survival and ANXA7 mRNA transcript expression (HR, 0.74; 95% CI, 0.57–0.95; P=.019).
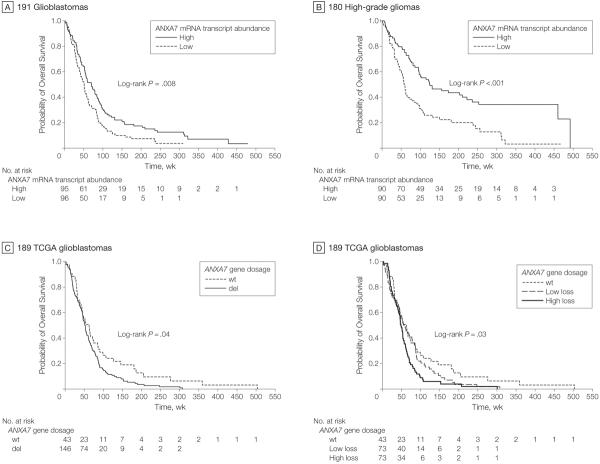
A 2-class model stratifying patients according to median ANXA7 mRNA transcript expression also demonstrated significance for ANXA7 (P=.008; log-rank test; HR, 0.667; 95% CI, 0.493–0.902), with median durations of survival of 70 vs 51 weeks and 46.9 vs 74.8 deaths per 100 person-years for high vs low ANXA7 expression, respectively (Figure 9A). Median duration of follow-up was 70 weeks (1.4, 479) and 51.1 weeks (range, 1.0–310.1) for the high and low ANXA7 mRNA transcript expression groups, respectively. Continuous ANXA7 mRNA transcript expression also demonstrated a significant association with survival in a unified cohort of 180 high-grade gliomas in a univariate Cox model (HR, 0.46; 95% CI, 0.31–0.70; P=.0002) as well as in a multivariate model including the clinically relevant covariates of patient age, O6-methylguanine-DNA methyltransferase, and tumor type (WHO classification) (HR, 0.66; 95% CI, 0.43–0.99; P=.046). A 2-class model based on median ANXA7 mRNA transcript expression confirmed a significant difference in survival between the high ANXA7 and low ANXA7 subgroups (log-rank P=.00003; HR, 0.476; 95% CI, 0.333–0.680), with median survival times of 125 vs 59 weeks and 21.8 vs 50.0 deaths per 100 person-years, respectively (Figure 9B). Median duration of follow-up was 118 weeks (range, 0.1–492) and 57.0 weeks (range, 1.0–467.0) for the high and low ANXA7 expression groups, respectively.
Figure 9. Association of ANXA7 and Survival of Patients With Malignant Glioma.
Kaplan-Meier estimates of overall survival in various glioblastoma/high-grade glioma populations according to annexin A7 (ANXA7) mRNA transcript expression or ANXA7 gene dosage. A, Survival estimates in 191 glioblastomas stratified into 2 classes based on median ANXA7 mRNA transcript abundance. Median follow-up for the groups of patients with high and low ANXA7 mRNA transcript expression was 70 weeks (range, 1.4–479) and 51.1 weeks (range, 1.0–310.1), respectively. B, Survival estimates in 180 high-grade gliomas stratified according to median ANXA7 mRNA transcript abundance. Median follow-up for the groups of patients with high and low ANXA7 mRNA transcript expression was 118 weeks (range, 0.1–492) and 57.0 weeks (range, 1.0–467.0), respectively. C, Survival estimates in 189 glioblastomas with available survival data from The Cancer Genome Atlas (TCGA) project analyzed for ANXA7 gene dosage and stratified according to ANXA7–wild-type (wt) vs ANXA7-deleted (del) status based on circular binary segmentation. Median follow-up for the groups of patients with ANXA7–wild-type and ANXA7-deleted status was 53.9 weeks (range, 1.1–503.4) and 50.8 weeks (range, 2.0–307.4), respectively. (D) Survival estimates in the same 189 TCGA glioblastomas grouped into 1 subgroup with ANXA7–wild-type status, and ANXA7-deleted tumors grouped into 2 equally sized subgroups of tumors with low-level (low loss) and high-level (high loss) gene dosage loss based on median gene dosage. Median follow-up was 53.9 weeks (range, 1.1–503.4), 52.9 weeks (range, 2.0–307.4), and 46.7 weeks (range, 2.9–300.6) for the ANXA7–wild-type, low-level and high-level ANXA7 loss groups, respectively.
ANXA7 Gene Dosage Matters Clinically
We assessed the association between ANXA7 gene dosage and patient outcome in 189 tumors of the TCGA project. Cox proportional hazards regression revealed a significant association with survival for ANXA7 continuous gene dosage (HR, 0.48; 95% CI, 0.28–0.83; P = .008), as did a 2-class model stratifying patients according to ANXA7 wild-type (n=43) vs deletion status (n=146) (log-rank P=.042; HR, 0.686; 95% CI, 0.476–0.989). Patients with wild-type ANXA7 vs ANXA7 deletion had a median survival of 63 weeks vs 51 weeks and 54.0 vs 80.1 deaths per 100 person-years, respectively (Figure 9C). Median duration of follow-up was 53.9 weeks (range, 1.1,–503.4) and 50.8 weeks (range, 2.0–307.4) for the ANXA7 wild-type and deletion groups, respectively. We also assigned the 189 tumors into 3 subgroups: one with wild-type ANXA7 and 2 equally sized subsets of ANXA7-deleted tumors distinguished as low-level allelic insufficiency (ie, low loss of functional gene dosage) and high-level allelic insufficiency relative to the median gene dosage of ANXA7-deleted tumors. We found this model to significantly relate to overall survival (log-rank P=.030; 54.0, 72.0, and 89.9 deaths/100 person-years for the ANXA7 wild-type, low-level, and high-level ANXA7 allelic insufficiency groups, respectively; Figure 9D), further supporting the importance of ANXA7 haploinsufficiency as an indicator of poor outcome in patients with glioblastomas. Median durations of follow-up were 53.9 weeks (range, 1.1–503.4), 52.9 weeks (range, 2.0–307.4), and 46.7 weeks (range, 2.9–300.6) for the ANXA7 wild-type, low-level, and high-level ANXA7 allelic insufficiency groups, respectively.
Chromosome 10q Residents ANXA7 and Phosphatase and Tensin Homolog (PTEN)
Because the PTEN (HGNC 9588) gene on 10q23.3 has been implicated as the key tumor suppressor gene on chromosome 10q,34,35 we analyzed whether the survival association of ANXA7 heterozygosity would be independent of PTEN status in the 189 tumors of the TCGA project. Since both genes colocalize physically on 10q (ANXA7, 10q21.1-q21.2; PTEN, 10q23.3) one would hypothesize mutual dependence. However, our gene dosage analysis in TCGA indicates small single-deletion events involving both genes.2 A multivariate Cox proportional hazard regression model, including continuous ANXA7 and PTEN gene dosage as covariates, revealed that ANXA7 gene dosage is independently associated with duration of patient survival (P = .008; HR, 0.33; 95% CI, 0.15–0.75), but PTEN gene dosage is not (P=.18; HR, 1.57; 95% CI, 0.81–3.03). Both univariate Cox proportional hazard regression (HR, 0.88; 95% CI, 0.59–1.30; P=.514) and a 2-class model based on deletion status (log-rank P=.106) failed to display a significant association between duration of survival and PTEN gene dosage. Finally, we compared a 2-class model that incorporates deletion information for both genes with the 2-class model based only on ANXA7 deletion status. We included in this model only those 177 tumors that either showed deletions of both ANXA7 and PTEN (n = 144) or showed wild-type status for both genes (n = 33). As opposed to the ANXA7 single-gene model, we did not find a significant difference in duration of survival between patients with ANXA7/PTEN codeleted tumors and those with ANXA7/PTEN wild-type tumors (log-rank P=.065). These analyses establish that an effect on ANXA7 gene dosage underlies the clinical relevance of loss of genetic material on chromosome 10q.
COMMENT
The dismal prognosis in glioblastoma outcome, even with the most advanced clinical care, addresses the need for the translation of new biological insights into clinical end points that can ultimately influence patient management. Identification of genes in which expression is altered or pathways in which activity is modified in tumors is important to understanding basic tumor biology, developing clinical-pathological correlations, and identifying points of therapeutic intervention. As we demonstrate here for ANXA7 and its link to EGFR signaling and dysregulation in glioblastomas, these require integration of genomic analysis, cancer genetics and biology, and clinical validation.
Dysregulation of EGFR signaling has been associated with a number of human cancers and represents the target of an expanding class of anticancer therapies. Gains and activating mutations of EGFR occur in about 45% of glioblastomas16 and are invariably associated with EGFR overexpression. About two-thirds of these tumors show overexpression of the corresponding protein, suggesting that additional mechanisms of EGFR deregulation exist.
Our integrated ANXA7 gene dosage and sequence analysis in TCGA and our functional data suggest that ANXA7 functions as a haploinsufficiency gene in glioblastomas. Although TCGA data indicate that glioblastomas harbor heterozygous (mono-allelic) ANXA7 losses and still express, yet at significantly diminished levels, the ANXA7 mRNA transcript from the retained allele, we found a seemingly low ANXA7 protein abundance in ANXA7-deleted tumors compared with that of wild-type tumors. This observation is consistent with previous findings in an ANXA7 haploinsufficiency model in mice, in which tumors formed spontaneously in a heterozygous (+/−) ANXA7 state show very low amounts of ANXA7 protein11 and suggests a significant though potentially nonlinear relationship between the ANXA7 gene and protein.
We propose that ANXA7 haploinsufficiency is a positive regulator of EGFR signaling and a driver for the conserved monosomy of chromosome 10 in glioblastomas. We provide evidence that ANXA7 loss of function facilitates unmitigated EGFR signaling, thereby contributing to an EGFR gain-of-function phenotype in high-grade gliomas, and that the complementary dysregulation of EGFR and ANXA7 synergistically promotes the tumorigenic potential of glioblastoma cells. Previous studies have reported a role of annexins—albeit not ANXA7—in mechanisms ensuring membrane translocation during signal transduction.36,37 We find the functional evidence to be further affirmed at the clinical level by revealing a significant negative relationship between duration of survival and loss of ANXA7 mRNA transcript expression in glioblastoma patients. We further show that the extent of allelic insufficiency (Box) for ANXA7 might be clinically relevant: in the analysis of crude tumor tissues, such as those in TCGA, the degree of allelic insufficiency describes an average characteristic of more or less heterogeneous cell populations in which certain tumor cells may harbor heterozygous ANXA7 deletions and others do not. Our data indicate that such average gene dosage profiles may affect clinical patient outcome. Nonetheless, exploitation of this relationship for clinical benefit awaits further investigation. Although our studies show ANXA7 at both gene and gene expression levels to be associated with the duration of survival in multiple retrospectively analyzed patient populations, its translation into a clinical end point requires prospective validation of its survival relationship, possibly involving screens for the ANXA7 protein. Future studies should further elucidate the EGFR dependence of the tumor supressor role of ANXA7 and the possibility of additional mechanisms contributing to its tumor-suppressive effects in high-grade gliomas. For example, some evidence suggests that ANXA7 heterozygous mice show down-regulation of PTEN,38 which could augment the glioma-promoting function of ANXA7.
The complex biology of glioblastomas suggests that the selective advantage of retaining a conserved monosomy of chromosome 10 may derive not from a single gene-gene interaction, but rather from several gene relationships that converge along a common signaling pathway. In keeping with this notion, it is likely that multiple tumor suppressors reside on chromosome 10, of which several may contribute to the deregulation of oncogenes acting on critical signaling pathways. Our data show that, at the very least, cross talk between ANXA7, PTEN, and EGFR lead to constitutive activation of PI3K-AKT signaling, a central pathway of tumor cell survival and proliferation.2
The multidimensional study of genomic regions of frequent alteration has already born fruit in cancer research by identifying critical genes in which molecular targeting has yielded clinical responses.39 Similarly, lessons learned from the in-depth mechanistic exploration of the ability of cooperating genes to confer a biological advantage for tumor-specific aneuploidies could help identify clinically meaningful points of therapeutic intervention. Combination therapies targeting such cooperating genes could evade problems encountered by molecular therapeutics that target isolated genes: genes with a cooperative role in deregulating a certain signaling pathway could decisively determine the response to such a therapeutic agent. This has been shown for the EGFR tyrosine-kinase inhibitor erlotinib in glioblastoma patients, in which the status of the chromosome 10 resident PTEN, which acts downstream of EGFR on the PI3K-AKT pathway, modifies the biological response of the tumor.40 The role of ANXA7 as a potential determinant of EGFR-targeted therapeutics should be explored further.
CONCLUSIONS
Our work provides a mechanistic explanation for the coselection of EGFR gains and monosomy of chromosome 10 in glioblastomas: allelic insufficiency of the chromosome 10 resident ANXA7 functions as a positive regulator of EGFR signaling. The biological synergy of deregulating tumorigenic-signaling pathways through coselected genetic mechanisms may significantly affect the success of our treatments: the genes conferring the biological advantage of such coselection may mutually determine or modify the biological response of a cancer to targeted therapeutics.
Box. Glossary.
Alleles are mutually exclusive alternative forms of the same gene occupying the same locus on homologous chromosomes, differing in DNA sequence and governing the same biochemical and developmental process. In diploid organisms with 2 copies of each chromosome, including humans, 2 alleles make up the individual's genotype.
Allelic insufficiency refers to a biologically relevant alteration in gene dosage caused by the loss of a functional allele.
Aneuploidy is deviation of the chromosomal constitution of cells from the normal by the addition or subtraction of chromosomes or chromosome pairs. In a normal diploid cell, the loss of a chromosome pair is termed nullisomy, the addition of a chromosome pair is tetrasomy, the loss of a single chromosome is monosomy, and the addition of a single chromosome is trisomy.
Double minute chromosomes are representative small circular fragments of extra-chromosomal DNA observed in many human tumors that frequently harbor amplified oncogenes and genes involved in drug resistance, therefore giving tumor cells selective advantage for growth and survival. Double minutes, like normal chromosomes, are composed of chromatin and replicate in the nucleus of the cell during cell division; but unlike typical chromosomes, they contain no centromere or telomere.
Gene knockdown is any of various techniques to reduce the expression of a gene without eliminating it entirely. The reduced expression may be permanent or via a transient mechanism.
Haploinsufficiency occurs when a diploid cell only has a single functional copy of a gene that does not produce enough of a gene product (typically a protein) to permit the cell to function normally, leading to an abnormal or diseased state.
Interactome is the whole set of molecular interactions in cells, most notably protein-protein interactions and protein-DNA interactions.
Monosomy is a chromosomal abnormality consisting of the absence of 1 chromo-some from the normal diploid number.
Definitions are in part adapted from the National Cancer Institute Terminology Browser (http://nciterms.nci.nih.gov/NCIBrowser/Dictionary.do) using National Cancer Institute thesaurus terminology.
Supplementary Material
Acknowledgments
Additional Contributions: We thank G. Dimri, PhD, NorthShore University HealthSystem Research Institute, Evanston, Illinois, for donating pIK packaging vector used in cotransfection of human shANXA7 knockdown retroviral construct in a pSM2 vector targeting the 1073–1093 mRNA sequence of ANXA7 and C. Svendsen, University of Wisconsin, Madison, for donating fetal neural stem cells (line Ctx M031) used in the pyrosequencing analysis. Neither of these individuals received any compensation in association with their contribution/gift.
Funding/Support: This work was supported by the State of Illinois Excellence in Academic Medicine Program (EAM Award 211 to Dr M. Bredel), by NIH/NCI RO1CA108633 and funds from The Ohio State University-James Comprehensive Cancer Center (both to Dr Chakravarti), by RTOG U10CA21661 and by CCOP U10CA37422. The results published here are in part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov.
Footnotes
Role of the Sponsor: The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Financial Disclosures: None reported.
Additional Information: The eFigure is available at http://www.jama.com.
REFERENCES
- 1.Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2003;2(3):202–210. [PubMed] [Google Scholar]
- 2.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302(3):261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furge LL, Chen K, Cohen S. Annexin VII and annexin XI are tyrosine phosphorylated in peroxovana-date-treated dogs and in platelet-derived growth factor-treated rat vascular smooth muscle cells. J Biol Chem. 1999;274(47):33504–33509. doi: 10.1074/jbc.274.47.33504. [DOI] [PubMed] [Google Scholar]
- 4.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6(6):449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 5.Shirvan A, Srivastava M, Wang MG, et al. Divergent structure of the human synexin (annexin VII) gene and assignment to chromosome 10. Biochemistry. 1994;33(22):6888–6901. doi: 10.1021/bi00188a019. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava M, Bubendorf L, Srikantan V, et al. ANX7, a candidate tumor suppressor gene for prostate cancer. Proc Natl Acad Sci U S A. 2001;98(8):4575–4580. doi: 10.1073/pnas.071055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leighton X, Srikantan V, Pollard HB, Sukumar S, Srivastava M. Significant allelic loss of ANX7region (10q21) in hormone receptor negative breast carcinomas. Cancer Lett. 2004;210(2):239–244. doi: 10.1016/j.canlet.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava M, Bubendorf L, Raffeld M, et al. Prognostic impact of ANX7-GTPase in metastatic and HER2-negative breast cancer patients. Clin Cancer Res. 2004;10(7):2344–2350. doi: 10.1158/1078-0432.ccr-03-0278. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava M, Torosyan Y, Raffeld M, Eidelman O, Pollard HB, Bubendorf L. ANXA7 expression represents hormone-relevant tumor suppression in different cancers. Int J Cancer. 2007;121(12):2628–2636. doi: 10.1002/ijc.23008. [DOI] [PubMed] [Google Scholar]
- 10.Torosyan Y, Dobi A, Naga S, et al. Distinct effects of annexin A7 and p53 on arachidonate lipoxygenation in prostate cancer cells involve 5-lipoxygenase transcription. Cancer Res. 2006;66(19):9609–9616. doi: 10.1158/0008-5472.CAN-06-1574. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava M, Montagna C, Leighton X, et al. Haploinsufficiency of Anx7 tumor suppressor gene and consequent genomic instability promotes tumorigenesis in the Anx7(+/−) mouse. Proc Natl Acad Sci U S A. 2003;100(24):14287–14292. doi: 10.1073/pnas.2235927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutschbauer AM, Jaramillo DF, Proctor M, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169(4):1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smilenov LB. Tumor development: haploinsufficiency and local network assembly. Cancer Lett. 2006;240(1):17–28. doi: 10.1016/j.canlet.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways [published online ahead of print September 4, 2008] Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Scheck AC, Cloughesy TF, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1(1):52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 20.Wright LS, Prowse KR, Wallace K, Linskens MH, Svendsen CN. Human progenitor cells isolated from the developing cortex undergo decreased neurogenesis and eventual senescence following expansion in vitro. Exp Cell Res. 2006;312(11):2107–2120. doi: 10.1016/j.yexcr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Pfisterer WK, Hank NC, Preul MC, et al. Diagnostic and prognostic significance of genetic regional heterogeneity in meningiomas. Neuro Oncol. 2004;6(4):290–299. doi: 10.1215/S1152851704000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvius M, Paulsson J, Weibrecht I, et al. In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol Cell Proteomics. 2007;6(9):1500–1509. doi: 10.1074/mcp.M700166-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Söderberg O, Gullberg M, Jarvius M, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 24.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281(5375):363, 365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 25.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20(1):273–285. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme [published online ahead of print September 4, 2008] Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark AJ, Ishii S, Richert N, Merlino GT, Pastan I. Epidermal growth factor regulates the expression of its own receptor. Proc Natl Acad Sci U S A. 1985;82(24):8374–8378. doi: 10.1073/pnas.82.24.8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Earp HS, Austin KS, Blaisdell J, et al. Epidermal growth factor (EGF) stimulates EGF receptor synthesis. J Biol Chem. 1986;261(11):4777–4780. [PubMed] [Google Scholar]
- 31.Jinno Y, Merlino GT, Pastan I. A novel effect of EGF on mRNA stability. Nucleic Acids Res. 1988;16(11):4957–4966. doi: 10.1093/nar/16.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin V, Mazzucchelli L, Frattini M. An overview of the epidermal growth factor receptor fluorescence in situ hybridisation challenge in tumour pathology. J Clin Pathol. 2009;62(4):314–324. doi: 10.1136/jcp.2008.059592. [DOI] [PubMed] [Google Scholar]
- 33.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 35.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 36.White IJ, Bailey LM, Aghakhani MR, Moss SE, Futter CE. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25(1):1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan RO, Martin-Almedina S, Garcia M, Jhoncon-Kooyip J, Fernandez MP. Deciphering function and mechanism of calcium-binding proteins from their evolutionary imprints. Biochim Biophys Acta. 2006;1763(11):1238–1249. doi: 10.1016/j.bbamcr.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava M, Eidelman O, Leighton X, Glasman M, Goping G, Pollard HB. Anx7 is required for nutritional control of gene expression in mouse pancreatic islets of Langerhans. Mol Med. 2002;8(12):781–797. [PMC free article] [PubMed] [Google Scholar]
- 39.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452(7187):553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.