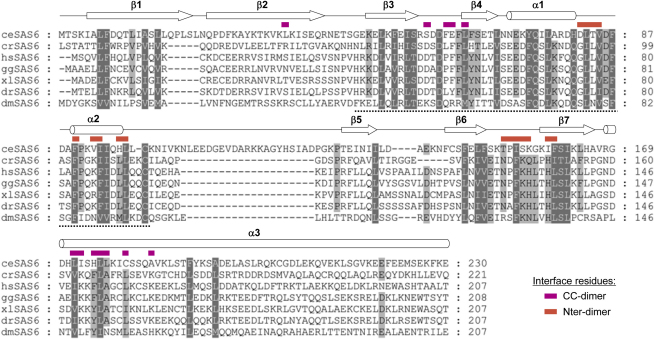
Figure S2.
Structure-Based Sequence Alignment of SAS-6 Orthologs, Related to Figure 2 and Figure 4
Highly conserved and conserved residues are highlighted in dark and light gray, respectively. Secondary structure assignments based on the crystal structures of C. elegans SAS-6 and Bld12p (Figures 2 and 4) are shown on top of the alignment. Interacting residues seen in the CC-dimer are indicated in magenta; the ones seen in the N-dimer are indicated in red. The PISA domain characteristic of SAS-6 proteins is indicated by a dashed black line at the bottom of the alignment. Species identifiers are: ce, Caenorhabditis elegans; hs, Homo sapiens; gg, Gallus gallus; xl, Xenopus laevis; dr, Dario rerio; dm, Drosophila melanogaster; cr, Chlamydomonas reinhardtii. UniProtKB/Swiss-Prot sequence accession identifiers are as follows: ceSAS-6, SAS6_CAEEL; Bld12p (crSAS-6), A9CQL4_CHLRE; hsSAS-6, SAS6_HUMAN; ggSAS-6, SAS6_CHICK; xlSAS-6, SAS6_XENLA; drSAS-6, SAS6_DANRE; dmSAS-6, SAS6_DROME.