Figure S4.
Characterization of C. reinhardtii Bld12p, Related to Figure 4
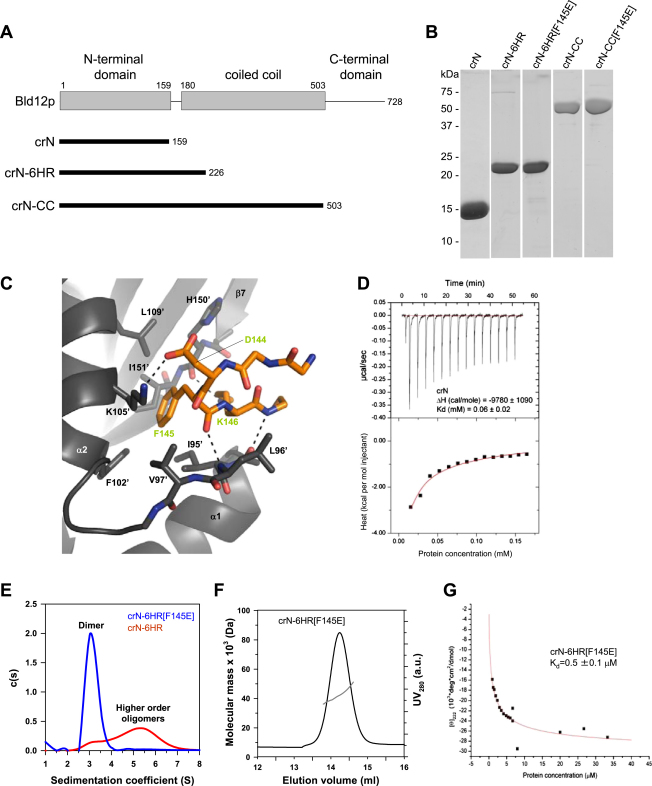
(A) Schematic representation of Bld12p and fragments generated in this study. crN, N-terminal domain; crN-6HR, N-terminal domains extended by 6 heptad repeats of the adjacent coiled coil; crN-CC; N-terminal domains extended by the full adjacent coiled coil. Numbers correspond to Bld12p amino acids.
(B) Coomassie-stained SDS-PAGE sections showing final purification products of the indicated recombinant proteins. Approximate molecular weights from in-gel markers are shown.
(C) Close up view of the interaction network seen at the crN-dimer interface in cartoon (main chains) and stick (contacting residues) representations. Monomers A and B are colored in dark gray and orange, respectively.
(D) Dissociation isotherm obtained by ITC for crN. Top panel: raw data representing the response to injections of crN at high concentration into sample buffer. Bottom panel: integrated heat change (closed squares) and associated curve fit (red solid line).
(E) Sedimentation velocity analysis of the crN-6HR (red) and crN-6HR[F145E] (blue). Protein concentration was 150 μM for both samples. The peak labeled with ‘Dimer’ corresponds to a molecular weight of ∼50 kDa, which is consistent with the formation of dimers. The region of S values highlighted with ‘Higher order oligomers’ is indicative of higher order oligomer formation beyond dimers.
(F) MALS analysis of crN-6HR[F145E]. The UV absorbance profile of size exclusion chromatography (black line) is overlaid with the molecular weight estimation by multi-angle light scattering (gray line). The determined molecular weight of 48.3 kDa is consistent with the formation of a stable dimer. Molecular weight of the crN-6HR[F145E] monomer: 25.8 kDa.
(G) crN-6HR[F145E] dilution series monitored by CD at 222 nm. The red solid line represents the fit to the data (closed squares) using a monomer-dimer model.