Abstract
To gain new insights into the mechanism of soybean (Glycine max) resistance to the soybean cyst nematode (Heterodera glycines), we compared gene expression profiles of developing syncytia in soybean near-isogenic lines differing at Rhg1 (for resistance to Heterodera glycines), a major quantitative trait locus for resistance, by coupling laser capture microdissection with microarray analysis. Gene expression profiling revealed that 1,447 genes were differentially expressed between the two lines. Of these, 241 (16.8%) were stress- and defense-related genes. Several stress-related genes were up-regulated in the resistant line, including those encoding homologs of enzymes that lead to increased levels of reactive oxygen species and proteins associated with the unfolded protein response. These results indicate that syncytia induced in the resistant line are undergoing severe oxidative stress and imbalanced endoplasmic reticulum homeostasis, both of which likely contribute to the resistance reaction. Defense-related genes up-regulated within syncytia of the resistant line included those predominantly involved in apoptotic cell death, the plant hypersensitive response, and salicylic acid-mediated defense signaling; many of these genes were either partially suppressed or not induced to the same level by a virulent soybean cyst nematode population for successful nematode reproduction and development on the resistant line. Our study demonstrates that a network of molecular events take place during Rhg1-mediated resistance, leading to a highly complex defense response against a root pathogen.
Soybean cyst nematode (SCN; Heterodera glycines) is the most important pathogen of soybean (Glycine max). The average bushels of soybean lost to SCN on an annual basis in the United States during 2006 to 2009 was 128.6 million bushels, and this loss was valued at $1.286 billion (Koenning and Wrather, 2010). The success of this obligate sedentary endoparasite is solely dependent on its ability to establish a permanent feeding cell, called the syncytium, within the roots of soybean. Infective juveniles penetrate into the root and migrate toward the vasculature. Near the vasculature, each juvenile selects a single cell, which is modified to allow for the incorporation of adjacent cells through progressive cell wall dissolution to form a multinucleate syncytium. The nematode derives nutrients from the syncytium for its growth and reproduction.
The primary management practice for this pathogen is resistant soybean cultivars. Resistant cultivars have been developed by identifying SCN-resistant soybean germplasm from collections of plant introductions (PIs) and incorporating the trait through conventional breeding programs. Although several sources of resistance have been identified, only a few PIs have been used in breeding programs, due to undesirable traits associated with other resistance sources. The most predominant sources of resistance found in commercially available cultivars are derived from PI 88788, PI 54840 (Peking), and PI 437654. In all resistant cultivars, the infective juveniles are capable of penetrating into roots and can induce the formation of syncytia, but the syncytia become necrotic soon after establishment and the nematodes starve to death. Although necrosis is a common theme, the timing of necrosis and the degeneration of syncytia vary among resistant cultivars, depending on the source of resistance (Acedo et al., 1984). For example, in Peking, syncytial collapse is observed as early as 48 h post infection (Mahalingam and Skorupska, 1996), whereas the onset of the resistance response is much slower in PI 209332, with degeneration of the syncytia not occurring until 8 to 10 d post infection (Acedo et al., 1984).
Despite the extensive histological studies documenting the cellular changes associated with degenerating syncytia in soybean (Endo, 1965; Riggs et al., 1973; Acedo et al., 1984), very little is known about the molecular mechanisms underlying syncytium collapse. Research conducted in the past decade has identified a number of quantitative trait loci associated with SCN resistance (for review, see Concibido et al., 2004) in different PIs that serve as sources of resistance in breeding programs. Among these, two major quantitative trait loci are Rhg1 (for resistance to Heterodera glycines) on soybean chromosome 18 (formerly linkage group G) and Rhg4 on chromosome 8 (formerly linkage group A2). Rhg1 exhibits incomplete dominance and contributes to a significant portion of SCN resistance in most PIs tested, including PI 88788, PI 90763, PI 209332, and Peking (Concibido et al., 2004). In addition, Rhg1 is effective against a broad spectrum of SCN populations. Rhg4 is dominant and is required for full resistance to certain SCN populations in some (e.g. Peking, PI 437654) but not all (e.g. PI 209332, PI 88788) resistant sources (Brucker et al., 2005). Despite cytological and molecular mapping studies, the genes for SCN resistance have not been identified (Melito et al., 2010), and the mechanism of resistance on the molecular level has yet to be fully elucidated.
Microarray analyses have been carried out to study this plant-nematode interaction. Initial studies used whole soybean roots infected with SCN to assess transcriptional changes during a compatible interaction (Khan et al., 2004; Alkharouf et al., 2006; Ithal et al., 2007a; Klink et al., 2007a). However, due to the specialized nature of the interaction and the location of syncytia well within the root, it is very difficult to draw meaningful conclusions using whole roots to understand this pathosystem. Laser capture microdissection (LCM) of syncytial cells coupled with microarray analysis has been particularly useful in extending our understanding of the SCN-soybean interaction, as indicated by recently published studies (Klink et al., 2005; Ithal et al., 2007b). These studies have provided new insights into the underlying molecular events occurring during syncytium development. More recently, the same technology has been applied to study incompatible SCN-soybean interactions (Klink et al., 2007b, 2009, 2010). Two studies reported on a comparative microarray analysis of soybean genes induced in response to either a virulent or an avirulent SCN population on Peking (Klink et al., 2007b, 2009), demonstrating that soybean can differentiate between nematode populations prior to feeding cell establishment (Klink et al., 2007b). The same group also published a microarray study that examined the transcriptional changes occurring in syncytia induced by an avirulent SCN population on PI 88788 at three time points after infection (Klink et al., 2010). To our knowledge, there are no reports of a direct comparative analysis of syncytia gene expression profiles using near-isogenic lines (NILs) to identify transcripts regulated by specific soybean resistance genes. NILs have several advantages over PIs for comparative analyses of plant gene expression between resistant and susceptible soybean in response to SCN. Theoretically, NILs can share up to 98% of their genome, differing only in a region encompassing a trait of interest (Li et al., 2004); thus, NILs are powerful tools to study the effects of specific gene loci with reduced genetic background effects. Consequently, the use of NILs for molecular studies is becoming more prevalent. NILs have been used in a microarray analysis of iron-efficient and -inefficient cultivars of soybean (O’Rourke et al., 2009) and a wheat (Triticum aestivum) leaf rust resistance gene, Lr10 (Manickavelu et al., 2010). NILs also recently helped to identify the effects of the Arabidopsis (Arabidopsis thaliana) gene FLC on seed germination (Chiang et al., 2009).
To gain new insights into the cause of the aberrant syncytia development that occurs in resistant soybean in response to SCN infection, we analyzed gene expression in syncytia induced in soybean NILs differing at the Rhg1 locus (Mudge, 1999). LCM coupled with comparative microarray profiling of syncytia isolated from the resistant NIL (NIL-R) and susceptible NIL (NIL-S) resulted in the identification of 1,447 differentially expressed genes using a false discovery rate (FDR) set at 10%. Many of the genes induced in NIL-R are soybean homologs of genes known to play important roles in disease resistance responses of other plant species to various pathogens, including canonical resistance genes (e.g. coiled-coil nucleotide-binding Leu-rich repeat class of receptors [CC-NB-LRR] class of receptors), genes associated with the hypersensitive-like response (HR), apoptotic cell death, the salicylic acid (SA)-mediated resistance pathway, and several transcription factors with defense-related roles. These results were validated by syncytia-specific quantitative PCR (qPCR) time-course qPCR on infected whole root pieces, and promoter-GUS reporter experiments. Our study reveals that Rhg1 mediates a complex defense response within syncytia formed in resistant soybean plants, ultimately limiting the growth and development of the nematode.
RESULTS
Response of NILs to SCN
Soybean NILs, derived from a cross between the susceptible cv Evans and the resistant PI 209332, were chosen for these studies. These NILs are predicted to share 98% of their genome, differing at the major SCN resistance locus, Rhg1 (Mudge, 1999). NIL-S is susceptible and NIL-R is resistant to SCN inbred line PA3 (HG type 0). The Rhg1 allele in PI 209332 is likely similar to the Rhg1 allele in PI 88788, the source of resistance found in greater than 90% of commercially available SCN-resistant soybean cultivars. Field populations of SCN that can break PI 88788 resistance typically can break PI 209332 resistance, suggesting that these PIs share a similar type of resistance (Colgrove and Niblack, 2008). The delayed resistance response in PI 209332 and PI 88788 is thought to be due to the absence of the Rhg4 resistance allele, which is present in Peking, a cultivar that exhibits a rapid resistance response to SCN. Our experimental system takes advantage of this slow resistance response to characterize Rhg1-mediated differences in gene expression during syncytium formation.
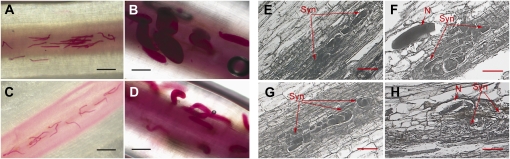
In laboratory inoculation assays, we studied the penetration and development of SCN and the formation of syncytia on the two NILs to identify appropriate time points for LCM analysis. Freshly hatched second-stage juveniles (J2s) were used for synchronized infection of soybean roots. Infected roots were harvested at different time points and stained with acid fuchsin to monitor the infection process. Similar numbers of nematodes were observed in both NIL-R and NIL-S at 2 d post inoculation (dpi; Fig. 1, A and C), indicating that resistance controlled by Rhg1 does not affect penetration and migration of nematodes in soybean roots. At 10 dpi, significant differences in the development of nematodes were observed. Late fourth-stage juveniles (J4) and early adult females were observed in the NIL-S (Fig. 1B) by 10 dpi, whereas the majority of the nematodes had only advanced to third-stage juveniles (J3) and early J4 in the NIL-R (Fig. 1D). These results are consistent with results reported by Li et al. (2004) in greenhouse bioassays using these NILs.
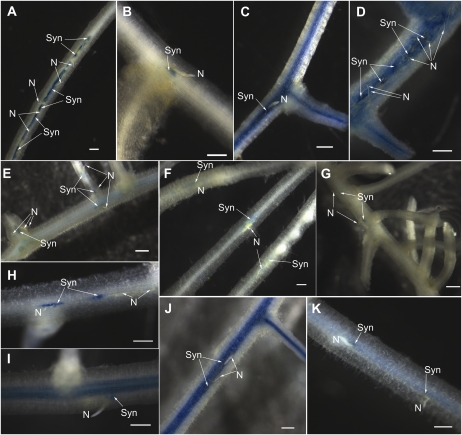
Figure 1.
Nematode development and syncytia formation on NILs of soybean. A to D, Penetration and development of SCN PA3 on resistant (NIL-R) and susceptible (NIL-S) lines. Roots of 2-d-old seedlings were infected with an equal number of SCN (PA3) juveniles, and the roots were acid fuschin stained at different dpi. A, NIL-S, 2 dpi. B, NIL-S, 10 dpi. C, NIL-R, 2 dpi. D, NIL-R, 10 dpi. E to H, Developmental differences between PA3-induced syncytia on NIL-S and NIL-R roots. E, NIL-S syncytium at 5 dpi. F, NIL-S syncytium at 8 dpi. G, NIL-R syncytium at 5 dpi. H, NIL-R syncytium at 8 dpi. N, Nematode; Syn, syncytia. Bars = 250 μm (A–D) and 50 μm (E–H).
To assess the differences in syncytium development at a more refined level, infected root samples at 5, 8, and 10 dpi were sectioned for microscopic examination. At 5 dpi, syncytia appeared normal in both the NIL-S (Fig. 1E) and NIL-R (Fig. 1G). Normal syncytium development was observed at 8 dpi in NIL-S (Fig. 1F), whereas degenerating cells both in and around developing syncytia were observed in NIL-R at 8 dpi (Fig. 1H). The majority of the syncytia were degenerated by 10 dpi in NIL-R. Based on these observations, we chose to assay gene expression changes within syncytia at 5 and 8 dpi in the NILs.
Transcript Profiling of Syncytia in NILs
The GeneChip Soybean Genome Array (Affymetrix), which carries 37,593 probe sets representing 35,611 soybean transcripts, was used to compare the transcriptional profiles of SCN-induced syncytia in NIL-R and NIL-S. The microarray analysis was carried out using copy RNA generated from LCM syncytia at 5 and 8 dpi with SCN from either the NIL-S or NIL-R. We found no significant evidence of interaction between NIL and dpi. Thus, we focus on the main effects of NIL and report differences between NIL-S and NIL-R that are averaged over 5 and 8 dpi. This comparison of expression profiles between genotypes resulted in the identification of 1,447 differentially expressed probe sets using a FDR set at 10%. We chose a FDR of 10% as the cutoff for significance to reduce the number of type 2 errors (i.e. truly differentially expressed genes that were not declared to be significantly differentially expressed). As discussed by Nettleton (2006), the number of type 2 errors can be quite large when FDR thresholds are kept too low. We felt that it was more important for our follow-up analyses to allow some additional type 1 errors (false discoveries) in order to reduce the number of type 2 errors.
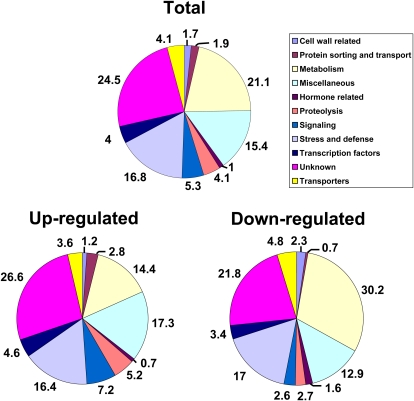
Of the 1,447 probe sets, 828 were up-regulated (Supplemental Table S1) and 619 were down-regulated (Supplemental Table S2) in the NIL-R compared with the NIL-S. The recently released SoyBase annotation (version 2) for the Affymetrix Soybean Genome Array was used to classify these genes into categories (Fig. 2). Of the 1,447 probe sets, 355 (24.5%) correspond to genes coding for unknown proteins and/or those with no known homologs in Arabidopsis, with a confidence value cutoff (E value < 10−6). Two other major categories include cellular metabolism genes (306 probe sets; 21.1%) and stress- and defense-related genes (241 probe sets; 16.8%). Additional classifications include (in descending order) cellular signaling, transporters, proteolysis, transcription factors, protein sorting and transport, cell wall-related, and hormone-related genes. Probe sets that do not fit into any of these categories or fall into multiple categories are grouped as “miscellaneous” (223; 15.4%). A number of probe sets corresponding to different soybean gene models had the same Arabidopsis homologs; this is not surprising given the duplicated nature of the soybean genome (Schlueter et al., 2004, 2007). These probe sets may represent homeologous genes with the same function, especially when their expression patterns fall within ±1-fold difference of each other.
Figure 2.
Functional classification of differentially expressed genes identified by microarray analysis.
qPCR Validation of Microarray Data
The microarray data were validated by qPCR analysis of selected genes using RNA isolated from syncytial cells laser microdissected from the roots of NIL-R and NIL-S at 5 dpi. The genes were selected to represent those both up- and down-regulated with fold changes ranging from 27.65-fold up-regulation to 17.63-fold down-regulation in the microarray analysis (Table I). Of the 42 genes tested, 38 genes (90.5%) showed differential expression in the same direction as that observed in the microarray experiment (Table I). Only four probe sets that showed a down-regulation in the microarray (Gma.2139.2.S1_S_at, GmaAffx.78421.1.S1_at, GmaAffx 84808.1.S1_at, and Gma.3504.2.S1_at; Table I) were slightly up-regulated in the qPCR analysis. Thus, overall, the qPCR results agreed with the microarray results.
Table I. qPCR validation of microarray results.
Values shown are fold change compared with NIL-S/PA3 at 5 dpi. Regular text indicates genes up-regulated; boldface text indicates genes down-regulated; underlined text indicates genes suppressed by the virulent TN19 SCN population; italicized text indicates genes not suppressed by the virulent TN19 SCN population. Asterisks indicate genes whose fold change in expression was opposite in qPCR and microarray.
| AffyChip Probe Set Identifier | Gene Model/EST Sequence | Putative Function | qPCR Fold Change: NIL-R/PA3 5 dpi | Microarray Fold Change: NIL-R/PA3 5 dpi | qPCR Fold Change: NIL-R/TN19 5 dpi |
| Gma.10240.1.A1_at | BE057471 | No predicted gene model | −23.44 | −2.05 | −128.82 |
| Gma.10701.2.S1_at | Glyma05g27030.1 | P450 pseudogene-like, mandelate racemase N-terminal domain | −1.86 | −4.85 | −2.63 |
| Gma.10796.1.S1_a_at | Glyma15g43040.1 | Cellulose synthase, CEV1-like | 2.95 | 1.34 | 1.15 |
| Gma.11004.1.S1_at | Glyma03g35920.1 | Harpin-induced family protein (YLS9)/HIN1 family protein | 15.49 | 8.09 | 2.45 |
| Gma.11334.1.S1_a_at | BI967327 | Caffeoyl-CoA 3-O-methyltransferase, putative | −1.82 | −2.76 | −2.63 |
| Gma.1423.1.S1_a_at | Glyma01g38450.1 | Brassinosteroid signaling positive regulator-related | 2.57 | 1.65 | 1.91 |
| Gma.14272.1.S1_at | Glyma10g28300.1 | Hydrophobic protein (RCI2B)/low temperature- and salt-responsive protein (LTI6B) | 33.88 | 11.69 | 2.09 |
| Gma.1445.1.S1_at | Glyma13g44700.1 | Cinnamoyl-CoA reductase, putative | −1.58 | −3.21 | −1.20 |
| Gma.15972.1.A1_at | Glyma04g17710.1 | Calcium-binding EF hand family protein | 16.60 | 27.65 | Not tested |
| Gma.16613.1.S1_s_at | Glyma10g40400.1 | Zinc finger (C2H2 type) family protein | 9.55 | 4.67 | 2.00 |
| Gma.16807.1.S1_at | Glyma09g14880.1 | Zinc finger (B-box type) family protein | 4.37 | 2.68 | 2.40 |
| Gma.17843.1.S1_at | Glyma04g02660.1 | Gibberellin-regulated protein 1 (GASA1)/gibberellin-responsive protein 1 | 16.98 | 2.62 | 2.75 |
| Gma.2139.2.S1_s_at | Glyma13g41960.1 | PfkB-type carbohydrate kinase family protein | 1.15* | −1.62 | 1.78 |
| Gma.2821.1.S1_at | Glyma01g42660.1 | Osmotin-like protein (OSM34) | 19.95 | 6.41 | −2.09 |
| Gma.3504.2.S1_at | Glyma13g00380.1 | WRKY family transcription factor | 1.48* | −2.59 | 1.95 |
| Gma.3990.1.S1_s_at | Glyma08g14370.1 | Ethylene-responsive calmodulin-binding protein | 2.09 | 1.9 | 7.76 |
| Gma.4071.1.S1_at | Glyma17g10050.2 | Gibberellin-regulated protein 2 (GASA2) | −2.75 | −5.26 | −6.17 |
| Gma.4207.1.S1_at | Glyma06g00630.1 | Myb family transcription factor (MYB32) | −1.23 | −2.15 | 1.78 |
| Gma.4565.1.S1_s_at | Glyma19g01590.2 | Gibberellin-regulated protein 3 (GASA3) | −1.38 | −4.76 | −20.89 |
| Gma.4589.1.S1_s_at | Glyma11g35800.1 | Senescence-associated protein, SAG20 | 4.07 | 2.46 | 1.45 |
| Gma.5283.1.S1_at | Glyma06g07300.1 | Plant natriuretic peptide | −2.19 | −17.63 | −724.44 |
| Gma.596.1.S1_at | Glyma13g34580.4 | 14-3-3 protein (GRF9) | −1.12 | −2.96 | −3.80 |
| Gma.6062.1.S1_at | Glyma03g32130.1 | Dehydration-responsive protein-related | −1.26 | −2.69 | 1.45 |
| Gma.7381.1.S1_at | Glyma20g33430.1 | NAC domain-containing protein | 2.45 | 1.67 | 2.34 |
| Gma.7526.1.A1_at | Glyma11g11430.1 | Senescence/dehydration-associated protein-related (ERD7) | 16.22 | 7.62 | 3.98 |
| Gma.7737.1.S1_at | BG650195 | MATE efflux family protein | 467.74 | 14.73 | 10.96 |
| Gma.8586.1.S1_at | Glyma17g17330.2 | Pro-rich family protein | 6.92 | 3.39 | 6.61 |
| Gma.8859.1.A1_at | Glyma15g03110.1 | Hyp-rich glycoprotein family protein | 21.38 | 5.17 | 3.55 |
| Gma.9553.1.A1_at | Glyma14g06080.1 | DREB subfamily A-2 of ERF/AP2 transcription factor family | 22.91 | 8.08 | 8.13 |
| GmaAffx.11502.1.S1_at | Glyma16g01640.1 | Pectinesterase family protein | 5.89 | 2.13 | 2.29 |
| GmaAffx.21079.1.A1_at | Glyma03g32400.1 | Plasmodesmal protein | 2.95 | 2.15 | 2.04 |
| GmaAffx.2744.1.S1_at | Glyma04g40930.1 | Auxin-responsive family protein | 6.31 | 3.53 | 2.88 |
| GmaAffx.29929.1.S1_at | Glyma20g29410.1 | DREB subfamily A-1 of ERF/AP2 transcription factor family (CBF3) | 15.49 | 11.25 | 3.63 |
| GmaAffx.3568.1.S1_at | Glyma02g19870.1 | bZIP transcription factor family protein (bZIP60) | 5.89 | 3.21 | 2.57 |
| GmaAffx.46603.1.S1_at | Glyma19g27260.1 | GRAM domain-containing protein/ABA-responsive protein-related | 15.14 | 5.55 | 2.29 |
| GmaAffx.494.1.S1_at | Glyma16g28970.2 | Chitinase A (CHIA) | 6.92 | 4.99 | 10.00 |
| GmaAffx.70008.1.S1_at | BU762337 | Myb family transcription factor (MYB20) | −131.83 | −3.89 | −630.96 |
| GmaAffx.74588.1.S1_at | Glyma12g34210.1 | Non-race-specific disease resistance protein, NDR1-like | 10.23 | 3.81 | 3.24 |
| GmaAffx.78421.1.S1_at | Glyma08g15650.1 | Pectinesterase family protein | 1.45* | −1.58 | −1.66 |
| GmaAffx.84808.1.S1_at | Glyma13g20810.2 | Ethylene-insensitive 2 (EIN2) | 1.91* | −1.59 | 1.82 |
| GmaAffx.88182.1.S1_at | Glyma03g35930.1 | Harpin-induced family protein/HIN1 family protein | 25.70 | 8.42 | 7.08 |
| GmaAffx.89435.1.A1_s_at | Glyma03g38190.2 | S-Adenosyl-Met synthetase 2 (SAM2) | 2.24 | 1.77 | 1.32 |
A comparative qPCR analysis for these genes was also carried out using RNA isolated from syncytial cells laser microdissected from soybean roots of the NIL-R infected with a virulent SCN population (TN19; HG type 1-7) at 5 dpi. Interestingly, a comparison between qPCR results of syncytia induced in the NIL-R by the virulent and avirulent (PA3; HG type 0) SCN populations showed that the extent of up-regulation or down-regulation of 35 (85.4%) of the 41 genes tested within syncytia induced by the virulent SCN was less than that attained by the avirulent population (underlined in Table I). These data indicate that the expression of many of these genes is either partially suppressed or not induced to the same level by the virulent SCN population for successful nematode reproduction and development on the NIL-R.
qPCR Analysis of Infected Whole Root Pieces
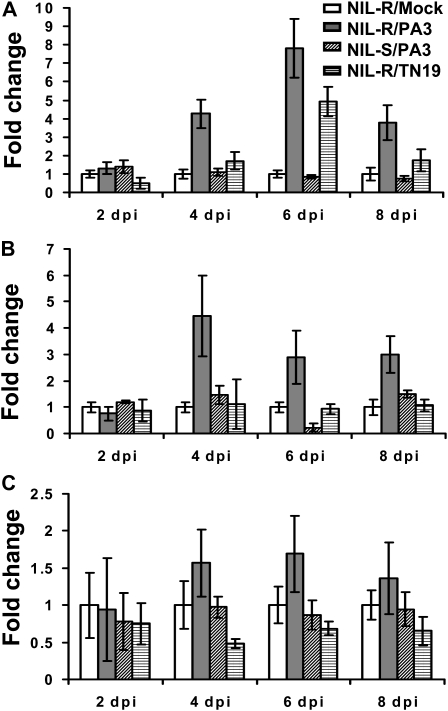
For purposes of microarray validation and to obtain a detailed temporal expression pattern of select differentially expressed genes, we conducted a qPCR analysis for three genes using RNA isolated from excised SCN-infected whole root pieces at different time points post inoculation. The NIL-R and NIL-S were mock inoculated or infected with either the avirulent (PA3) or virulent (TN19) SCN population, and root pieces were excised from infection sites at 2, 4, 6, and 8 dpi. Root pieces from 10 different roots were bulked for each sample, and total RNA was isolated for qPCR analysis. Time courses of expression were conducted for three different genes (represented by probe sets Gma.7623.1.A1_at, 87.9-fold; GmaAffx.68498.1.S1_at, 13.1-fold; and GmaAffx.46603.1.S1_at, 5.6-fold; Supplemental Table S1). The expression of mock-inoculated NIL-R at each time point was set as 1. At 2 dpi, the expression of all three genes was more or less equal between treatments (Fig. 3). However, by 4 dpi, differential expression was observed for all three genes between the NIL-S and NIL-R. Up-regulation is very clear in the case of genes highly up-regulated in the microarray (Fig. 3, A and B) and barely detectable in the case of GmaAffx.46603.1.S1_at (Fig. 3C), which had the lowest fold up-regulation based on microarray analysis. This clearly illustrates the dilution effect attributed to using infected whole root pieces for gene expression analyses in this pathosystem. For Gma.7623.1.A1_at, up-regulation in the NIL-R peaks at 6 dpi (Fig. 3A). The pattern is similar for GmaAffx.46603.1.S1_at (Fig. 3C). The expression pattern is slightly different for the gene represented by probe set GmaAffx.68498.1.S1_at in that the maximum up-regulation is observed at 4 dpi. These peaks in expression levels are consistent with the timing of the resistance response observed in syncytia of the NIL-R (Fig. 1). The observed trend in expression for each gene over the time course of infection was reproducible in three independent infection experiments; however, the level of up-regulation varied among experiments. This difference can be attributed to the biological variation inherent to nematode infection experiments, where it is impossible to achieve identical rates of infection. Similar to what was observed in qPCR analyses of RNA isolated from syncytia, infection by the virulent and avirulent SCN populations results in differential up-regulation of these genes. For all three genes tested, the level of up-regulation is lower in response to infection by the virulent nematode population (Fig. 3). Additionally, in contrast to infection of the NIL-R, infection of the NIL-S with the avirulent population shows only a slight up-regulation for all three genes tested.
Figure 3.
qPCR analysis of up-regulated genes in excised infected whole root pieces of resistant (NIL-R) and susceptible (NIL-S) NILs at different dpi with avirulent (PA3) or virulent (TN19) SCN. Comparison of gene expression levels was made with the NIL-R mock-infected roots (taken as 1) at the indicated time points post inoculation. A, Gma.7623.1.A1_at. B, GmaAffx.68498.1.S1_at. C, GmaAffx.46603.1.S1_at. The qPCR results are normalized to a soybean ubiquitin (accession no. D28123) endogenous control. The graph is representative of three independent experiments, and the bars represent the difference between minimum and maximum relative quantification values, which are calculated from the sd of Ct values using ABI software.
Promoter-GUS Expression Analysis
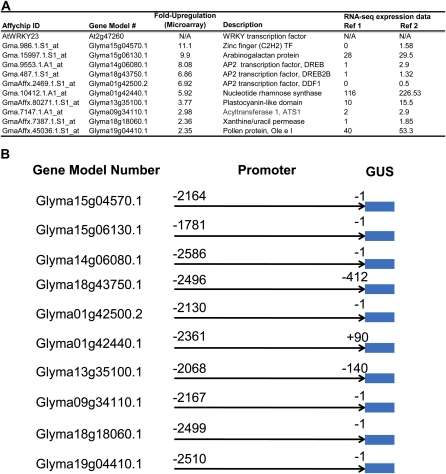
Promoter-GUS fusions were generated to provide further validation of the spatial expression pattern of the differentially expressed genes identified by microarray analysis and to isolate nematode-responsive soybean promoter sequences with high levels of expression within syncytia. For these experiments, primers corresponding to the 5′ upstream sequences of 10 genes (Fig. 4A), chosen from the differentially expressed microarray data set (Supplemental Table S1), were designed by using the recently released Williams 82 soybean genome sequence (Schmutz et al., 2010). Genes selected for promoter analysis were those whose expression was highly up-regulated based on our microarray analysis or genes presumed to either not be expressed or expressed weakly in roots based on expression data of Arabidopsis orthologs, because at the time we performed this experiment, we did not have reliable expression data for these genes in the roots of soybean. Promoter fragments were amplified by PCR using Williams 82 genomic DNA as a template and cloned upstream of a GUS reporter gene in the Gateway binary vector pYXT1 (Xiao et al., 2005; Fig. 4B). Transgenic soybean hairy roots were generated in the NIL-R soybean background for each reporter construct. As a positive control, the Arabidopsis WRKY23 promoter (At2g47260) was tested in soybean; At2g47260 is induced within syncytia in Arabidopsis upon infection with the beet (Beta vulgaris) cyst nematode, Heterodera schachtii (Grunewald et al., 2008). The transgenic hairy roots were infected with the avirulent (PA3) SCN population. The positive control and all 10 promoter-GUS lines show induced GUS gene expression at the nematode feeding sites at 5 dpi (Fig. 5). For several promoter-GUS lines, GUS expression is observed throughout the root but further induced at nematode feeding sites (Fig. 5, C, D, I, and J; Supplemental Fig. S1, B and C). Several promoters have an expression pattern that is very low or more restricted to specific cell types within roots, and an up-regulation of GUS expression is clearly distinguishable at the nematode feeding sites (Fig. 5, A, B, E–H, and K; Supplemental Fig. S1, A and D). RNA-seq expression data for each soybean gene model in uninfected roots according to Severin et al. (2010) and Libault et al. (2010) are provided in Figure 5A for comparison. Although there are some discrepancies (Fig. 5, D, G, and I), our promoter-GUS expression pattern in roots largely conforms to the RNA-seq data.
Figure 4.
Description of promoter-GUS reporter constructs used for microarray validation. A, Description of probe sets with soybean gene models used for promoter isolation and their putative function; RNA-seq expression data for soybean gene models in uninfected roots is according to Severin et al., 2010 (Ref 1) and Libault et al., 2010 (Ref 2). B, Schematic showing the lengths of promoter elements cloned and their coordinates with respect to the soybean gene model. [See online article for color version of this figure.]
Figure 5.
Promoter-GUS expression in transgenic soybean hairy root lines of resistant NIL-R infected with PA3 SCN. Promoter-GUS constructs representing 10 genes up-regulated in NIL-R identified from the microarray analysis and the nematode-inducible AtWRKY23 were infected with SCN and stained for GUS expression at 5 dpi. A, AtWRKY23 (At2g47260). B, Glyma15g04570.1. C, Glyma15g06130.1. D, Glyma14g06080.1. E, Glyma09g34110.1. F, Glyma01g42500.2. G, Glyma01g42440.1. H, Glyma18g43750.1. I, Glyma18g18060.1. J, Glyma19g04410.1. K, Glyma13g35100.1. Images are representative of at least five independent hairy root lines for each promoter-GUS fusion construct. N, Nematode; Syn, syncytia. Bars = 500 μm.
Differential Expression of Stress- and Defense-Related Genes
We specifically looked at stress- and defense-related genes to gain a better understanding of the Rhg1-associated syncytium collapse that occurs in the NIL-R in response to SCN. The differential expression of this class of genes varies from 87-fold up-regulated to 17-fold down-regulated (Tables II and III; Supplemental Tables S1 and S2). Soybean orthologs of many known plant defense genes have not yet been identified; therefore, we relied on their similarity to Arabidopsis homologs. A total of 241 probe sets representing 16.8% of the total number of differentially expressed genes identified are classified in this group. These included genes involved in apoptosis and disease resistance. Several soybean genes showing high similarity to defense genes of Arabidopsis that play a role in incompatible responses to other plant pathogens were found to be differentially expressed in syncytia of the NIL-R in response to SCN (Tables II and III; Supplemental Tables S1 and S2). These include a highly up-regulated AtBag6 homolog, a CC-NB-LRR gene, several heat shock protein (HSP) genes and heat shock transcription factors (HSFs), defense-related WRKY transcription factors, pathogenesis-related (PR) genes, and genes that modulate SA and jasmonic acid (JA) levels. A large number of genes involved in oxidative, drought, cold, osmotic, and salt stress responses are also differentially regulated (Tables II and III; Supplemental Tables S1 and S2).
Table II. Up-regulated stress- and defense-related genes.
| AffyChip Probe Set Identifier | q Value | Fold Change | Gene Model | At No. | Description |
| Gma.7623.1.A1_at | 0.0313 | 87.891 | Glyma07g06750.1 | AT2G46240.1 | Similar to AtBAG6 |
| GmaAffx.75438.1.S1_at | 0.0641 | 62.307 | Glyma17g13820.1 | AT1G76650.1 | Similar to Arabidopsis CML38 |
| GmaAffx.25859.1.S1_at | 0.0764 | 30.214 | Glyma14g06910.1 | AT3G46230.1 | Class I small heat shock protein (sHSP) family |
| GmaAffx.39393.1.S1_at | 0.0347 | 16.162 | Glyma10g32000.1 | AT4G10250.1 | Endomembrane-localized small heat shock protein |
| GmaAffx.91194.1.S1_at | 0.0376 | 14.499 | Glyma01g41920.1 | AT4G17260.1 | Putative l-lactate/malate dehydrogenase |
| Gma.17947.1.S1_at | 0.0565 | 13.481 | Glyma08g07340.1 | AT2G29500.1 | Small heat shock protein (HSP17.6B, class I) |
| Gma.14272.1.S1_at | 0.0396 | 11.694 | Glyma10g28300.1 | AT3G05890.1 | Rare cold-inducible 2B (RCI2B) |
| Gma.10763.1.S1_at | 0.0664 | 11.364 | Glyma04g05720.1 | AT5G12020.1 | Class II heat shock protein (HSP17.6, class II) |
| GmaAffx.29929.1.S1_at | 0.0268 | 11.25 | Glyma20g29410.1 | AT4G25480.1 | DREB subfamily A-1 of ERF/AP2 transcription factor family (CBF3) |
| Gma.2044.1.S1_at | 0.0722 | 10.771 | Glyma09g31740.1 | AT5G66400.2 | ABA- and drought-induced Gly-rich dehydrin protein |
| GmaAffx.68621.1.A1_at | 0.0988 | 9.439 | Glyma20g29770.1 | AT5G52300.2 | Induced in response to water deprivation |
| GmaAffx.88182.1.S1_at | 0.0448 | 8.423 | Glyma03g35930.1 | AT2G35980.1 | NDR1 and harpin-like (NHL) gene |
| Gma.11004.1.S1_at | 0.036 | 8.094 | Glyma03g35920.1 | AT2G35980.1 | NDR1 and harpin-like (NHL) gene |
| Gma.9553.1.A1_at | 0.0317 | 8.079 | Glyma14g06080.1 | AT2G40340.1 | DREB subfamily A-2 of ERF/AP2 transcription factor family |
| Gma.5637.1.S1_at | 0.0595 | 7.885 | Glyma10g28610.1 | AT2G47180.1 | Arabidopsis galactinol synthase 1 (AtGolS1) |
| Gma.7526.1.A1_at | 0.0312 | 7.621 | Glyma11g11430.1 | AT2G17840.1 | Drought-inducible gene |
| GmaAffx.63418.1.S1_at | 0.0568 | 7.081 | Glyma09g16810.1 | AT5G06720.1 | Endomembrane-located putative peroxidase |
| Gma.2821.1.S1_at | 0.0362 | 6.407 | Glyma01g42660.1 | AT4G11650.1 | Osmotin-like protein, soybean PR5 |
| GmaAffx.30428.1.S1_at | 0.0996 | 6.375 | Glyma17g08020.1 | AT1G16030.1 | Heat shock protein 70B (HSP70b) |
| Gma.8386.1.S1_at | 0.0268 | 6.08 | Glyma04g37040.1 | AT1G76650.1 | Similar to Arabidopsis CML38 |
| GmaAffx.6438.1.S1_at | 0.0678 | 5.926 | Glyma11g29720.1 | AT2G38470.1 | Similar to WRKY33 |
| GmaAffx.92590.1.S1_at | 0.0766 | 5.848 | Glyma02g33070.1 | AT2G41680.1 | NADPH thioredoxin sulfide reductase |
| Gma.13195.1.S1_s_at | 0.0274 | 5.445 | Glyma01g20860.1 | AT1G13340.1 | Unknown protein; oxidative stress |
| GmaAffx.6519.1.S1_at | 0.0624 | 5.408 | Glyma08g29470.1 | AT1G13340.1 | Unknown protein; oxidative stress |
| Gma.1622.1.A1_s_at | 0.0273 | 5.246 | Glyma05g17470.1 | AT5G66900.1 | Putative disease resistance protein (CC-NBS-LRR class) |
| Gma.5820.1.S1_s_at | 0.0268 | 4.856 | Glyma06g19810.1 | AT1G19020.1 | Unknown protein; oxidative stress |
| Gma.1439.1.S1_at | 0.0317 | 4.217 | Glyma04g14800.3 | AT3G22370.1 | Alternative oxidase AOX1a |
| GmaAffx.71308.2.A1_at | 0.0337 | 4.003 | Glyma04g05500.2 | AT2G26150.1 | Similar to AtHSF-A2 |
| Gma.620.1.S1_at | 0.0661 | 3.931 | Glyma15g40190.1 | AT1G78380.1 | Glutathione transferase of τ-GST gene family |
| GmaAffx.74588.1.S1_at | 0.0398 | 3.806 | Glyma12g34210.1 | AT3G20600.1 | NDR1-like |
| Gma.4089.1.S1_at | 0.0276 | 3.722 | No soybean match | AT4G25200.1 | AtHSP23.6-mito mRNA |
| GmaAffx.36259.1.S1_s_at | 0.0581 | 3.493 | Glyma05g33200.1 | AT2G02120.1 | Plant defensin (PDF) family PR protein |
| Gma.8204.1.A1_at | 0.0771 | 3.462 | Glyma04g14800.3 | AT3G22370.1 | Alternative oxidase AOX1a |
| Gma.8458.1.S1_at | 0.0922 | 3.265 | Glyma08g20190.1 | AT1G55020.1 | Lipoxygenase, similar to AtLOX1 |
| GmaAffx.3568.1.S1_at | 0.0196 | 3.208 | Glyma02g19870.1 | AT1G42990.1 | Similar to AtbZIP60 |
| GmaAffx.92499.1.S1_s_at | 0.0613 | 3.123 | Glyma19g43460.1 | AT3G04720.1 | Similar to antifungal chitin-binding protein hevein, PR-4 |
| GmaAffx.80951.1.S1_at | 0.0682 | 3.042 | Glyma16g29750.1 | AT5G52640.1 | AtHSP90.1 homolog |
| GmaAffx.84566.1.S1_x_at | 0.0539 | 3.011 | Glyma18g49360.1 | AT3G28910.1 | Transcription factor AtMYB30 homolog |
| Gma.7922.1.A1_a_at | 0.0431 | 2.88 | Glyma05g02210.1 | AT5G62520.1 | Similarity to RCD1 but without the WWE domain |
| GmaAffx.92919.1.S1_at | 0.0878 | 2.674 | Glyma05g05290.1 | AT5G47120.1 | Encodes BI-1, a homolog of mammalian Bax inhibitor 1 |
| GmaAffx.93596.1.S1_at | 0.040 | 2.672 | Glyma17g03950.2 | AT5G49520.1 | Similar to WRKY48 |
| GmaAffx.34450.1.S1_at | 0.0734 | 2.669 | Glyma05g05290.1 | AT5G47120.1 | Encodes BI-1, a homolog of mammalian Bax inhibitor 1 |
| GmaAffx.19934.1.S1_at | 0.0416 | 2.593 | Glyma13g21490.2 | AT5G03720.1 | Heat stress transcription factor HSFA3 |
| GmaAffx.1338.1.S1_at | 0.0962 | 2.555 | Glyma02g35210.1 | AT3G11820.1 | Similar to SYP121(PENETRATION1/PEN1) |
| Gma.8336.1.S1_at | 0.047 | 2.398 | Glyma19g40560.1 | AT2G47260.1 | Similar to WRKY23 |
| Gma.4639.1.A1_at | 0.0963 | 2.281 | Glyma13g36340.1 | AT3G20600.1 | NDR1-like |
| GmaAffx.22821.1.S1_at | 0.0687 | 2.178 | Glyma19g44390.2 | AT3G03300.2 | Encodes a Dicer-like 2 |
| Gma.11115.2.S1_at | 0.0268 | 2.146 | Glyma11g14950.1 | AT3G12580.1 | Heat shock protein 70 (HSP70) |
| GmaAffx.11781.1.S1_s_at | 0.0273 | 2.137 | Glyma02g35660.1 | AT5G06320.1 | NDR1 and harpin-like (NHL) gene |
| GmaAffx.89654.1.A1_s_at | 0.0338 | 2.063 | No soybean match | AT1G21750.1 | Protein disulfide isomerase-like (PDIL) protein |
| GmaAffx.20155.1.S1_at | 0.0517 | 2.036 | Glyma02g35230.1 | AT3G11820.1 | Similar to SYP121(PENETRATION1/PEN1) |
| Gma.10639.1.S1_x_at | 0.065 | 1.793 | Glyma03g27560.1 | AT2G41010.1 | Similar to a calmodulin-binding protein |
| Gma.6474.1.A1_s_at | 0.0655 | 1.727 | Glyma15g13470.1 | AT2G34690.1 | Similar to ACD11 gene |
| GmaAffx.1991.1.S1_at | 0.0637 | 1.665 | Glyma11g04040.2 | AT5G47120.1 | Encodes BI-1, a homolog of mammalian Bax inhibitor 1 |
| GmaAffx.80733.1.S1_at | 0.0396 | 1.578 | Glyma17g11240.1 | AT5G20320.1 | Encodes a Dicer-like 4 |
| Gma.3755.1.S1_at | 0.0869 | 1.481 | Glyma03g30590.1 | AT5G13320.1 | Similar to PBS3/GH3.12 |
| GmaAffx.19777.1.A1_at | 0.0986 | 1.43 | No soybean match | AT4G31800.2 | Similar to WRKY18 pathogen-induced transcription factor |
| GmaAffx.48022.2.A1_at | 0.0658 | 1.429 | Glyma13g02470.3 | AT4G08500.1 | Encodes MEKK1, phosphorylates MEK1 |
Table III. Down-regulated stress- and defense-related genes.
| AffyChip Probe Set Identifier | q Value | Fold Change | Gene Model | At No. | Description |
| Gma.5283.1.S1_at | 0.037 | −17.624 | Glyma06g07300.1 | AT2G18660.1 | Plant natriuretic peptide A (PNP-A) |
| Gma.5629.2.S1_a_at | 0.081 | −5.461 | Glyma15g05820.1 | AT2G41480.1 | Peroxidase, response to oxidative stress |
| GmaAffx.84317.1.S1_at | 0.054 | −5.457 | Glyma06g42310.1 | AT5G54250.2 | Similar to AtCNGC4, HR |
| Gma.5629.1.S1_at | 0.032 | −5.021 | No soybean match | AT2G41480.1 | Peroxidase, response to oxidative stress |
| Gma.5971.1.S1_at | 0.047 | −4.813 | Glyma11g07670.1 | AT5G66390.1 | Similar to Arabidopsis PER72, oxidative stress |
| GmaAffx.74124.1.S1_at | 0.083 | −3.937 | No soybean match | AT5G67400.1 | Similar to PER73 (RHS19), oxidative stress |
| Gma.1539.1.S1_at | 0.064 | −3.677 | Glyma11g05300.2 | AT4G37520.1 | Peroxidase similar to PER50, oxidative stress |
| GmaAffx.73002.1.S1_at | 0.060 | −3.570 | Glyma02g09470.1 | AT1G14790.1 | Similar to RDRP |
| Gma.4126.1.S1_at | 0.058 | −3.461 | Glyma06g16810.1 | AT5G63660.1 | Similar to defensin PDF2.5 defense response |
| Gma.4919.1.S1_at | 0.027 | −3.198 | Glyma14g05840.1 | AT5G05340.1 | Peroxidase, response to oxidative stress |
| GmaAffx.92030.1.S1_at | 0.095 | −2.594 | Glyma19g44310.1 | AT2G46370.2 | Similar to JAR1 |
| Gma.3504.2.S1_at | 0.050 | −2.591 | Glyma13g00380.1 | AT4G31550.1 | Similar to WRKY11 |
| GmaAffx.65280.1.A1_at | 0.078 | −2.574 | Glyma13g26640.2 | AT3G27890.1 | NADPH quinone oxidoreductase |
| Gma.8020.3.S1_at | 0.074 | −2.521 | Glyma13g07490.1 | AT3G25780.1 | Similar to AOC3 |
| Gma.338.1.S1_at | 0.027 | −2.438 | Glyma07g39020.1 | AT4G21960.1 | Peroxidase, response to oxidative stress |
| Gma.1502.1.S1_at | 0.087 | −2.285 | Glyma05g29400.1 | AT2G29420.1 | Similar to Arabidopsis GST τ-7 |
| Gma.2350.1.S1_at | 0.054 | −2.227 | No soybean match | AT5G60640.1 | PDIL-4 homolog, oxidative stress |
| Gma.8020.2.S1_a_at | 0.084 | −2.112 | No soybean match | AT1G13280.1 | Similar to AOC4, jasmonic acid biosynthesis |
| Gma.4207.1.S1_at | 0.027 | −2.147 | Glyma06g00630.1 | AT4G34990.1 | AtMYB32 homolog |
| GmaAffx.6478.1.S1_s_at | 0.068 | −2.051 | Glyma13g00380.1 | AT4G31550.1 | Similar to WRKY11 |
| Gma.3504.1.S1_at | 0.095 | −1.965 | Glyma17g06450.1 | AT4G31550.1 | Similar to WRKY11 |
| Gma.4189.1.S1_at | 0.054 | −1.942 | Glyma17g01720.1 | AT4G21960.1 | PRXR1, oxidative stress |
| Gma.3504.2.S1_a_at | 0.079 | −1.923 | Glyma13g00380.1 | AT4G31550.1 | Similar to WRKY11 |
| GmaAffx.54278.1.S1_at | 0.078 | −1.782 | Glyma13g03600.1 | AT1G21750.1 | Similar to PDIL1-1, regulation of programmed cell death |
| Gma.2677.1.S1_s_at | 0.064 | −1.731 | Glyma19g44310.1 | AT2G46370.2 | Similar to AtAR1, production of JA-Ile |
| Gma.2749.1.S1_at | 0.048 | −1.699 | Glyma10g40140.1 | AT1G80600.1 | Similar to WIN1, defense response |
| Gma.2350.1.S1_s_at | 0.097 | −1.645 | Glyma13g40130.1 | AT5G60640.1 | PDI1-4, oxidative stress |
| Gma.4312.1.S1_at | 0.098 | −1.633 | Glyma08g05200.1 | AT2G31570.1 | Glutathione peroxidase 2 (GPX2) |
| GmaAffx.89649.1.S1_s_at | 0.052 | −1.631 | Glyma05g34490.4 | AT2G43350.1 | ATGPX3 |
| GmaAffx.84808.1.S1_at | 0.039 | −1.590 | Glyma13g20810.2 | AT5G03280.1 | Ethylene-insensitive 2 (EIN2) |
| GmaAffx.85352.1.S1_at | 0.091 | −1.542 | Glyma03g33850.1 | AT5G03280.1 | Ethylene-insensitive 2 (EIN2) |
| Gma.4312.1.S1_x_at | 0.097 | −1.461 | Glyma08g05200.1 | AT2G31570.1 | Glutathione peroxidase 2 (GPX2) |
| Gma.3301.1.S1_at | 0.093 | −1.426 | Glyma06g00440.1 | AT4G02600.2 | Homology to MLO1 protein, cell death defense response |
| GmaAffx.90444.1.S1_s_at | 0.058 | −1.323 | Glyma11g21260.1 | AT3G27890.1 | NADPH quinone oxidoreductase |
DISCUSSION
The planting of resistant soybean cultivars is the primary strategy used to manage SCN population levels in the field. Despite the widespread use of SCN-resistant soybean, this pathogen still causes an estimated $1.286 billion annually in yield losses. A lack of understanding of the molecular basis of resistance to this pathogen continues to hinder progress to enhance the effectiveness and durability of natural plant resistance and enable the design of novel strategies for resistance through biotechnological approaches. Rhg1, a major resistance locus in almost all SCN-resistant germplasm, is required for resistance against multiple SCN H. glycines types (Concibido et al., 2004); however, the molecular nature of the resistance gene underlying Rhg1 (Melito et al., 2010) and the downstream signaling and response genes mediated by Rhg1 are not known. The Rhg1 gene has been mapped to chromosome 18 and is within 0.4 cm of simple sequence repeat marker satt_309 (Cregan et al., 1999), enabling the generation of NILs differing only at this locus (Mudge, 1999). These NILs are useful for molecular studies to dissect the SCN-soybean incompatible interaction because of the multigenic nature of resistance. Therefore, to develop a better understanding of the molecular events associated with Rhg1-mediated resistance against SCN, we employed the use of NILs for a comparative analysis of syncytial gene expression using LCM and microarrays. These NILs have been used previously to study the effects of Rhg1 on root penetration and development of SCN (Li et al., 2004). Although root penetration by SCN juveniles is similar between NIL-R and NIL-S, the growth, development, and fecundity of nematode females is suppressed on NIL-R (Li et al., 2004), suggesting that Rhg1 may have a negative impact on syncytium development and maintenance. The histological characteristics of syncytia in resistant soybean cultivars to SCN infection are well documented (Ross, 1958; Endo, 1965; Riggs et al., 1973; Acedo et al., 1984). Second-stage SCN juveniles (J2s) induce the formation of syncytia in all resistant cultivars, but syncytial collapse occurs several days later. In one type of resistance (PI 437654; Peking type), syncytial collapse is very rapid and begins to occur within 48 h of induction of the syncytium (Mahalingam and Skorupska, 1996). The NILs used in our study, however, show a delayed type of resistance (Acedo et al., 1984; Li et al., 2004), with notable histological changes to syncytia occurring by 8 to 10 dpi (Fig. 1). Thus, 5- and 8-dpi time points were chosen for laser capture of syncytia to reflect gene expression prior to the onset of syncytium collapse. The comparison of syncytia gene expression between NIL-R and NIL-S by microarray analysis identified 1,447 differentially expressed probe sets. We found a high representation of stress- and defense-related genes (241 probe sets representing 16.8% of the total; Fig. 2; Supplemental Tables S1 and S2), including genes involved in oxidative, heat, cold, salt, and drought stress. Due to space constraints, we limited our analysis to stress- and defense-related genes.
A gene coding for a BCL2-associated athanogene (BAG) domain protein with highest homology to the Arabidopsis BAG6 protein (AtBAG6) is the most highly up-regulated gene in syncytia of the resistant line (87-fold; Table II). Bag6 encodes a stress-induced calmodulin-binding BAG domain protein that is homologous to mammalian BAG proteins, which are regulators of BCL2 involved in apoptosis (Kang et al., 2006). BAG proteins are antiapoptotic in animals; however, overexpression of AtBag6 in yeast and Arabidopsis causes cell death (Kang et al., 2006). The increased expression of this gene in the resistant line suggests that the syncytia may be undergoing an apoptosis-like cell death response.
Several HSPs of the small HSP superfamily are up-regulated in syncytia of the resistant line (Table II). HSPs are stress-responsive proteins that have a protective function in promoting cellular stress tolerance (Wang et al., 2004). Small HSPs bind and stabilize denatured proteins to which other high-Mr HSPs act as chaperones under stress conditions. Several other HSPs, including an HSP70 homolog (Gma.11115.2.S1_at), HSP70B homolog (GmaAffx.30428.1.S1_at), HSP90.1 homolog (GmaAffx.80951.1.S1_at), and two HSFs, Hsf-A2 homolog (GmaAffx.71308.2.A1_at, 4.0-fold) and Hsf-A3 homolog (GmaAffx.19934.1.S1_at, 2.6-fold), are up-regulated in syncytia of the NIL-R. HSP90 is a highly conserved molecular chaperone rapidly induced during pathogen challenge and a variety of environmental stresses. It interacts with the R protein, RPM1 (Hubert et al., 2003), and is required for RPS2-mediated resistance against Pseudomonas syringae pv tomato DC 3000 (avrRpt2) (Takahashi et al., 2003). HSFs are involved in a variety of environmental stresses; Hsf-A2, for example, is a key inducer of defense responses and is up-regulated during environmental stress and hydrogen peroxide treatment (Nishizawa et al., 2006). AtBAG6 is up-regulated by heat stress, and the Hsf-A2 is involved in its regulation (Nishizawa et al., 2006).
Several genes related to endoplasmic reticulum (ER) stress were also found to be up-regulated (e.g. BZIP60 homolog [GmaAffx.3568.1.S1_at], BIP2 homolog [Gma.17631.1.S1_at], calnexin [Gma.6427.2.S1_a_at], Bax inhibitor genes [GmaAffx.1991.1.S1_at, GmaAffx.34450.1.S1_at, and GmaAffx.92919.1.S1_at], and several protein disulfide isomerases; Table II; Supplemental Table S1). ER stress is a cellular condition in which unfolded proteins accumulate in the ER. Misfolding of proteins is a result of mutations, disturbances in calcium homeostasis, and the heightened need for protein folding. In order to maintain ER homeostasis under such conditions, signaling pathways are activated that are collectively known as the unfolded protein response. When ER stress is not relieved by different measures, an apoptotic cell death occurs (Urade, 2009). Recently, it was reported that water deficit or drought leads to programmed cell death mediated by the ER stress response pathway in Arabidopsis roots (Duan et al., 2010). Several drought- and abscisic acid (ABA)-induced genes were found to be up-regulated in syncytia of the resistant NIL. Taken together, these data suggest that multiple stress response pathways are induced by an upstream signaling event in the resistant plants during nematode infection that may ultimately lead to the activation of an HR-like programmed cell death, causing the pathogen to starve and die. It is also possible that pathogen death occurs before the syncytial HR-like programmed cell death. The HR may represent the final stages of the resistance response, where a certain threshold of defense-related responses has been reached (Morel and Dangl, 1997). For example, the Arabidopsis dnd1 mutant expresses resistance to pathogens that otherwise induce HR in wild-type plants (Clough et al., 2000).
The production of reactive oxygen species (ROS) is a key aspect of the HR during R-mediated resistance to other pathogens (Lamb and Dixon, 1997). An NADPH thioredoxin reductase, similar to Arabidopsis NTRC, is up-regulated 5.8-fold (GmaAffx.92590.1.S1_at). Alternative oxidase (Gma.1439.1.S1_at, 4.2-fold; Gma.8204.1.A1_at, 3.46-fold), glutathione S-transferase (GST; Gma.620.1.S1_at, 3.9-fold), and a gene similar to RCD1-5 involved in ROS regulation (Gma.7922.1.A1_a_at, 2.9-fold) are up-regulated. Several genes related to oxidative stress and the regulation of ROS are also down-regulated (Table III), as are many peroxidases (Gma.5629.2.S1_a_at, Gma.5629.1.S1_at, Gma.5971.1.S1_at, GmaAffx.74124.1.S1_at, Gma.1539.1.S1_at, Gma.4919.1.S1_at, Gma.338.1.S1_at, and Gma.4189.1.S1_at, fold changes ranging from −5.4 to −1.9). Peroxidases are involved in hydrogen peroxide catabolism, and their down-regulation may suggest a positive impact on ROS generation, although, they can also generate ROS species (Passardi et al., 2004). Other down-regulated oxidative stress genes include two NADPH quinone oxidoreductases (GmaAffx.65280.1.A1_at, −2.57-fold; GmaAffx.90444.1.S1_s_at, −1.3-fold), glutathione peroxidase 2 and 3 homologs, and a protein disulfide isomerase-like4 (PDIL-4) that belongs to the thioredoxin family.
A gene coding for a CC-NB-LRR protein is up-regulated (Gma.1622.1.A1_s_at, 5.2-fold). The up-regulation of a MAP3K homolog (GmaAffx.48022.2.A1_at) implicates mitogen-activated protein kinase signaling in the regulation of resistance to SCN. A soybean gene encoding a protein with homology to Arabidopsis syntaxin 121 (SYP121), a secretory pathway protein with known roles in defense responses, is also induced (GmaAffx.1338.1.S1_at, 2.6-fold; GmaAffx.20155.1.S1_at, 2-fold), as are several genes involved in cold, drought, dehydration, and ABA responses (Gma.14272.1.S1_at, 11.7-fold; Gma.2044.1.S1_at, 10.8-fold; GmaAffx.68621.1.A1_at, 9.4-fold; Gma.7526.1.A1_at, 7.6-fold) and two transcription factors of the AP2/ERF family involved in drought responses (GmaAffx.29929.1.S1_at, 11.2-fold; Gma.9553.1.A1_at, 8.1-fold). The high up-regulation of these genes may suggest new roles in HR against a biotrophic pathogen, or they are secondary physiological responses that potentiate HR.
The most highly down-regulated probe set (Gma.5283.1.S1_at, −17.6-fold) corresponds to a gene encoding a predicted natriuretic peptide with an expansin-like domain sharing homology with AtPNP-A (Table III), which is involved in plant growth and homeostasis (Morse et al., 2004). AtPNP-A is induced by SA and is expressed at higher levels in Arabidopsis mutants with increased SA levels (Meier et al., 2008). Another highly down-regulated gene is a cyclic nucleotide-gated channel (CNGC; GmaAffx.84317.1.S1_at, −5.5 fold; Supplemental Table S4), which shares homology with Arabidopsis CNGC4/HLM1. Arabidopsis mutants of CNGC4/HLM1 produce a lesion-mimic phenotype and altered HR (Balague et al., 2003).
Several PR genes are also up-regulated. A soybean osmotin (Gma.2821.1.S1_at; Table II), which is described as a salt stress-induced acidic isoform of PR-5 (Onishi et al., 2006) and has similarity to Arabidopsis osmotin 34, is up-regulated 6.4-fold. Osmotins are components of incompatible reactions against bacterial pathogens (Jia and Martin, 1999). Another up-regulated PR protein is a hevein-like protein belonging to the PR-4 family, which is up-regulated during salt stress, in response to viral infection, and in systemic acquired resistance (Potter et al., 1993). We found a 3.5-fold up-regulation of a defensin homologous to Arabidopsis defensin PDF2.1 (GmaAffx.36259.1.S1_s_at) but a 3.5-fold down-regulation of another member of the same defensin family, PDF2.5 (Gma.4126.1.S1_at; Table III).
WRKY transcription factors are known to take part in defense responses to viral, bacterial, and fungal pathogens (Eulgem and Somssich, 2007). Several WRKY transcription factor homologs are up-regulated in syncytia of the NIL-R (Table II), including a homolog of AtWRKY33, a known regulator of defense pathways mediating resistance to P. syringae and fungal necrotrophic pathogens (Zheng et al., 2006; GmaAffx.6438.1.S1_at, 5.9-fold), an AtWRKY48 homolog (GmaAffx.93596.1.S1_at, 2.67-fold), and a homolog of AtWRKY18, which is involved in SA-mediated defenses against viruses, bacteria, and fungi (GmaAffx.19777.1.A1_at, 1.4-fold). Interestingly, a soybean homolog of AtWRKY23 (Gma.8336.1.S1_at, 2.4-fold), which is involved in nematode feeding site establishment (Grunewald et al., 2008), is also up-regulated. Down-regulated WRKYs (Table III) include a homolog of AtWRKY11 (GmaAffx.6478.1.S1_s_at, −2.05-fold; Gma.3504.1.S1_at, −2-fold; Gma.3504.2.S1_a_at, −1.9-fold), a negative regulator of basal defense responses against bacterial pathogens (Journot-Catalino et al., 2006). Down-regulation of a negative regulator would lead to an enhanced defense response.
In general, the SA pathway has been shown to be activated in resistance against biotrophs and the JA pathway in resistance to necrotrophs and insects, although exceptions exist (Glazebrook, 2005; Bari and Jones, 2009). The SA pathway also has been implicated in resistance to the root-knot nematode in tomato (Solanum lycopersicum; Branch et al., 2004). Here, we identified several homologs of genes belonging to the SA-mediated defense signaling pathway to be up-regulated in SCN-induced syncytia of the NIL-R. A soybean MYB protein homologous to AtMYB30, an SA-dependent R2-R3 MYB that acts as a positive regulator of HR cell death and is a modulator of SA levels (Vailleau et al., 2002; Raffaele et al., 2006), is up-regulated 3-fold. Other genes included soybean homologs of Arabidopsis NDR1 and NDR1/HIN1-like (NHL) genes, which are key signal transducers in SA-mediated signaling. NDR1 is a plasma membrane-localized protein required for disease resistance to P. syringae pv tomato DC3000 carrying avirulence genes avrRpm1, avrRpt2, avrPph3, and avrB. It is also required for resistance against avirulent isolates of the fungal pathogen Peronospora parasitica (Century et al., 1995, 1997). The requirement for resistance against a diverse group of pathogens suggests that this is a common downstream element in R gene-mediated resistance in plants. Arabidopsis ndr1 mutants have reduced ROS production and SA accumulation in response to avirulent bacteria (Shapiro and Zhang, 2001). Conversely, overexpression of NDR1 in Arabidopsis leads to enhanced resistance to virulent P. syringae pv tomato (Coppinger et al., 2004). NHL proteins have sequence homology to NDR1 of Arabidopsis and HIN1 of tobacco and are pathogen induced in Arabidopsis (Varet et al., 2002; Zheng et al., 2004). NHL3 overexpression in Arabidopsis is associated with enhanced resistance to virulent strains of P. syringae (Varet et al., 2003). We also identified homologs of Arabidopsis ACD11 (Gma.6474.1.A1_s_at, 1.7-fold) and PBS3 (Gma.3755.1.S1_at, 1.5-fold), which are involved in SA-mediated defense. PBS3 (WIN3), which interacts with the P. syringae effector protein HopW1-1, is important for responses induced by several effectors in Arabidopsis. PBS3 is an important component of NDR1-dependent RPS2-mediated resistance against P. syringae pv tomato carrying avrRpt2 and also plays a role in basal resistance (Lee et al., 2007). PBS3 is an acyl adenylase, and the Arabidopsis pbs3 mutant exhibits enhanced susceptibility to P. syringae pv tomato carrying avrPphB (Nobuta et al., 2007). In the pbs3 mutant, induced free and conjugated SA levels are reduced. A homolog of another Arabidopsis gene related to SA accumulation, WIN1, is down-regulated in syncytia of NIL-R. Overexpression of WIN1 delays SA accumulation in response to several effectors, including HopW1-1 (Lee et al., 2008), which indicates that WIN1 is a negative regulator. Thus, down-regulation of this gene would be predicted to have a positive impact on SA levels.
Previously, we found a majority of JA pathway components to be suppressed during a compatible soybean-SCN interaction (Ithal et al., 2007b). In this study, we found a homolog of Arabidopsis lipoxygenase 1 (AtLOX1; Table II) to be up-regulated in syncytia of the NIL-R, possibly suggesting the involvement of lipid peroxides in the resistance response. The up-regulation of soybean LOX genes is also reported in syncytia induced on Peking and PI 88788 by an avirulent population of SCN (Klink et al., 2009, 2010). Lipoxygenases have a role in basal resistance to the root-knot nematode in maize (Zea mays; Gao et al., 2008). Recently, mutations in AtLOX1 and silencing of a homologous gene of Capsicum annuum (CaLOX1) have been shown to increase susceptibility to diverse microbial pathogens (Hwang and Hwang, 2010). CaLOX1-silenced plants show lowered SA and ROS levels. However, we also identified down-regulation of two soybean genes corresponding to homologs of allene oxide cyclases (AOCs) involved in JA biosynthesis and a homolog of JAR1, a protein required to convert JA to the biologically active JA-Ile. These discrepancies emphasize the need for further studies directed at silencing the genes involved in SA and JA biosynthesis and quantifying hormone levels in nematode-infected roots to clarify the role of these small molecules in SCN-induced resistance in soybean.
Here, we present evidence for the potential involvement of a complex stress- and defense-related response, including increased expression of genes involved in the production of ROS, the unfolded protein response, SA-mediated signaling, and plant programmed cell death in Rhg1-mediated resistance to SCN. Involvement of almost all hormones shows an intricate network of cross talk associated with this defense response. Our study also highlights the importance of conducting a direct comparison between syncytia transcriptomes in the resistant versus susceptible NILs, using the same nematode population to identify genes potentially involved in resistance. Inadvertently, a large number of genes would be overlooked in a direct comparison of syncytia transcriptomes induced in the resistant line by an avirulent versus a virulent SCN population. Here, we demonstrate that the plant still mounts a defense response against the virulent nematode population, albeit somewhat attenuated compared with the avirulent nematode population. In contrast, the response of the susceptible line to the avirulent population is minimal.
Additionally, our study led to the identification of nematode-inducible soybean promoters, several of which have restricted expression in roots but are highly up-regulated in syncytia, which can be employed as a tool for more targeted RNA silencing experiments of soybean genes in the resistant background. Our study, together with the newly developed functional analysis tools in soybean such as virus-induced gene silencing (Zhang et al., 2009, 2010) and the recently completed soybean genome sequence (Schmutz et al., 2010), will hasten research to understand this relatively unknown, but fascinating, belowground incompatible plant-pathogen interaction and may ultimately lead to the development of novel strategies to enhance the nematode resistance of crop plants.
MATERIALS AND METHODS
Plant and Nematode Material
Seeds of soybean (Glycine max) NIL differing at the Rhg1 locus (NIL-R and NIL-S) were derived from a cross between the resistance source PI 209332 and the susceptible cv Evans (Mudge, 1999). The SCN (Heterodera glycines ‘Ichinohe’) inbred populations PA3 and TN19 were obtained from a publicly available collection at the University of Illinois at Urbana-Champaign and mass selected according to standard procedures (Niblack et al., 1993) on soybean cv Williams 82 and PI 437654, respectively. HG type tests (Niblack et al., 2002) confirmed that the PA3 population was HG type 0 and the TN19 population was HG type 1-7.
LCM
PA3- and TN19-infected root pieces (approximately 1 cm) of the NIL-R and NIL-S were excised at 5 or 8 dpi and immediately processed for LCM according to Ithal et al. (2007b).
Microarray Hybridization, Statistical Analysis, and qPCR Validation
RNA extraction, amplification, and labeling were performed according to Ithal et al. (2007b). The samples were sent to the Iowa State University GeneChip microarray core facility for fragmentation, hybridization, staining, and scanning of the GeneChip Soybean Genome Array (Affymetrix). The logarithms of the Affymetrix MAS 5.0 signals were normalized by computing the median of the log signals on each chip and then aligning these medians to a common value. These normalized expression data were analyzed on a gene-by-gene basis using the statistical computing package SAS (SAS Institute, Inc.). Each analysis was based on a randomized complete block design with three replications as blocks and the four combinations of genotype (resistant versus susceptible) and times post infection (5 and 8 dpi) as treatments. Tests for genotype main effects, dpi main effects, and genotype-by-dpi interaction were conducted for each gene. The resulting P values were converted to q values as described by Storey and Tibshirani (2003). These q values were used to control the estimated FDR at desired levels. For example, by declaring differential expression between resistant and susceptible genotypes for all genes with q values less than or equal to 0.10, the proportion of false positives among all genes declared differentially expressed is expected to be approximately 10%. Annotations and classifications were based on the SoyBase Affymetrix GeneChip Soybean Genome Array Annotation, version 2 (http://soybase.org/AffyChip/). Arabidopsis (Arabidopsis thaliana) unique gene identifiers (At numbers) were downloaded from The Arabidopsis Information Resource (www.arabidopsis.org) for the top hits. The microarray data are deposited in the ArrayExpress database at the European Bioinformatics Institute under accession number E-MEXP-3073. qPCR validation studies were conducted according to Ithal et al. (2007b).
Promoter-Reporter Constructs
The promoter sequences for the genes used in the GUS reporter assays were identified and downloaded from the soybean genome database (Phytozome; www.phytozome.net). Primers (Supplemental Table S3) were designed to amplify an approximately 2-kb sequence immediately 5′ of the ATG start site (Fig. 4B). The promoter DNA fragments were PCR amplified using soybean cv Williams 82 genomic DNA as a template and cloned into the Gateway cloning vector pDONR-Zeo (Invitrogen). The cloned promoters were sequenced using vector-specific primers and internal sequencing primers. The correct promoter fragments were then Gateway cloned into a pYXT1 vector (Xiao et al., 2005) upstream of a GUS gene as a transcriptional fusion. The final plasmids were verified by PCR analysis and used for the transformation of Agrobacterium rhizogenes (strain K599).
Hairy Root Transformation
Hairy roots transgenic for each promoter-GUS construct were generated using the method described by Wang et al. (2007) with the following modifications. The cotyledons were excised from 9-d-old aseptically grown soybean seedlings (NIL-R or cv Williams 82) and vacuum infiltrated for 20 min with A. rhizogenes culture resuspended in quarter-strength Gamborg’s salt solution (Phytotechnology Lab) carrying various reporter constructs. Cotyledons were cocultivated with A. rhizogenes for 3 d. The cotyledons were later placed on MXB medium (1× Murashige and Skoog basal nutrient salts [Gibco BRL], 1× Gamborg’s vitamins, 3% [w/v] Suc, and 0.8% [w/v] Daishin agar, pH 5.7) supplemented with kanamycin (200 μg mL−1) and timentin (238 μg mL−1) and incubated in a growth chamber at 26°C set to a long-day photoperiod (16 h of light/8 h of dark). Hairy roots that emerged after 14 d were root-tip propagated twice on MXB medium with kanamycin (200 μg mL−1) and timentin (238 μg mL−1), after which the roots were transferred to MXB medium with timentin (237 μg mL−1). Hairy roots at this stage were either used immediately for nematode inoculation experiments or maintained by subculturing for later use.
Nematode Infection of Transgenic Hairy Roots and GUS Staining
Infective second-stage juveniles (J2) were hatched from eggs as described by Wang et al. (2007). Nematodes were surface sterilized with sterilizing solution (0.004% [w/v] mercuric chloride, 0.004% [w/v] sodium azide, and 0.002% [v/v] Triton X-100) for 8 min followed by five washes with sterile water and resuspended in 0.1% (w/v) agarose. Hairy roots (3–4 cm) grown on MXB medium were inoculated approximately 1 cm above the root tip with 200 ± 25 J2s per root in a 25-μL volume. The roots were cut and stained for GUS expression at 5 dpi. GUS staining was done according to Jefferson (1987). Briefly, hairy roots were cut 1 to 2 cm above the infection zone and placed in GUS staining solution (100 mm Tris, pH 7.0, 50 mm NaCl, 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 1.5 mm potassium ferricyanide, pH 7.0, and 0.06% [v/v] Triton X-100). The root tissues were vacuum infiltrated twice for 10 min each and incubated at 37°C overnight. The GUS staining reaction was stopped by replacing staining solution with 70% (v/v) ethanol. GUS-stained roots were photographed with a Leica MZFLIII stereoscope (Leica Microsystems) fitted with an Optronics MagnaFire, version 2.0, camera (Optronics). Sectioning was done according to Wang et al. (2007).
Sample Preparation for Time-Course qPCR Analysis
Infected root tissues for time-course qPCR analysis were prepared as described by Ithal et al. (2007a), except that samples were collected at 2, 4, 6, and 8 dpi. Excised root pieces from 12 to 15 different plants were pooled for each genotype/inoculum combination. Samples were quick frozen in liquid nitrogen and stored at −80°C until RNA isolation. Nematode penetration was verified by staining the nematodes in at least five sample roots for each treatment at 24 h post inoculation as described by Ithal et al. (2007a). Infected root tissues from three independent biological replicates were prepared.
RNA Isolation and qPCR
Total RNA was isolated from root tissues using the RNeasy plant miniprep kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA synthesis was carried out using a SuperScript III first-strand synthesis kit (Invitrogen) according to the manufacturer’s instructions. Real-time qPCR was carried out using an Applied Biosystems 7500 real-time PCR system. Gene-specific primers (Supplemental Table S4) were designed using Primer Express software (Applied Biosystems). All quantitative reverse transcription-PCR was carried out in triplicate. PCR was performed using the following cycling parameters: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. A soybean ubiquitin gene (accession no. D28123) was used as an endogenous control. We determined by quantitative reverse transcription-PCR that expression of this gene is stable across the treatment groups in our experiment. Expression was quantified using the ΔΔCT method in comparison with the endogenous control. Fold changes were determined relative to the NIL-R mock-inoculated sample for each time point. There were no significant expression differences between mock-treated NIL-S and NIL-R roots.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Longitudinal cross-sections of promoter-GUS-stained transgenic soybean hairy root lines in the NIL-R background infected with PA3 SCN.
Supplemental Table S1. Classification of up-regulated genes at FDR < 0.1.
Supplemental Table S2. Classification of down-regulated genes at FDR < 0.1.
Supplemental Table S3. Gateway cloning primers used to generate promoter-GUS reporter fusions.
Supplemental Table S4. qPCR primers used for microarray validation and time-course qPCR.
Acknowledgments
We thank Nevin Young and JoAnn Mudge for the generous gift of NIL-R and NIL-S seed, Dave Sleper for seed bulking, Terry Niblack for providing the original SCN inbred populations, and Chris Town for the pYXT1 vector. We thank Robert Heinz for maintenance of the nematode cultures, Xuejing Yang for assistance with the generation and maintenance of hairy root cultures, Greg Yeckel for technical assistance, the University of Missouri Histology Laboratory for tissue processing, and Melody Kroll for proofreading the manuscript.
References
- Acedo JR, Dropkin VH, Luedders VD. (1984) Nematode population attrition and histopathology of Heterodera glycines-soybean associations. J Nematol 16: 48–56 [PMC free article] [PubMed] [Google Scholar]
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF. (2006) Time course microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224: 838–852 [DOI] [PubMed] [Google Scholar]
- Balague C, Lin B, Alcon C, Flottes G, Malmstrom S, Kohler C, Neuhaus G, Pelletier G, Gaymard F, Roby D. (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Branch C, Hwang CF, Navarre DA, Williamson VM. (2004) Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol Plant Microbe Interact 17: 351–356 [DOI] [PubMed] [Google Scholar]
- Brucker E, Carlson S, Wright E, Niblack T, Diers B. (2005) Rhg1 alleles from soybean PI 437654 and PI 88788 respond differentially to isolates of Heterodera glycines in the greenhouse. Theor Appl Genet 111: 44–49 [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K. (2009) Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr, Bent AF. (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgrove AL, Niblack TL. (2008) Correlation of female indices from virulence assays on inbred lines and field populations of Heterodera glycines. J Nematol 40: 39–45 [PMC free article] [PubMed] [Google Scholar]
- Concibido VC, Diers BW, Arelli PR. (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44: 1121–1131 [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ. (2004) Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J 40: 225–237 [DOI] [PubMed] [Google Scholar]
- Cregan PB, Mudge J, Fickus EW, Danesh D, Denny R, Young ND. (1999) Two simple sequence repeat markers to select for soybean cyst nematode resistance conditioned by the rhg1 locus. Theor Appl Genet 99: 811–818 [Google Scholar]
- Duan Y, Zhang W, Li B, Wang Y, Li K, Sodmergen Han C, Zhang Y, Li X. (2010) An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol 186: 681–695 [DOI] [PubMed] [Google Scholar]
- Endo BY. (1965) Histological responses of resistant and susceptible soybean varieties, and backcross progeny to entry development of Heterodera glycines. Phytopathology 55: 375–381 [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Gao X, Starr J, Gobel C, Engelberth J, Feussner I, Tumlinson J, Kolomiets M. (2008) Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol Plant Microbe Interact 21: 98–109 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inze D, Beeckman T, Gheysen G. (2008) A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol 148: 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK. (2010) The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol 152: 948–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Hearne L, Maier T, Baum TJ, Mitchum MG. (2007a) Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol Plant Microbe Interact 20: 293–305 [DOI] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG. (2007b) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol Plant Microbe Interact 20: 510–525 [DOI] [PubMed] [Google Scholar]
- Jia Y, Martin GB. (1999) Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease-resistant tomato plants. Plant Mol Biol 40: 455–465 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CH, Jung WY, Kang YH, Kim JY, Kim DG, Jeong JC, Baek DW, Jin JB, Lee JY, Kim MO, et al. (2006) AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ 13: 84–95 [DOI] [PubMed] [Google Scholar]
- Khan R, Alkharouf N, Beard H, Macdonald M, Chouikha I, Meyer S, Grefenstette J, Knap H, Matthews B. (2004) Microarray analysis of gene expression in soybean roots susceptible to the soybean cyst nematode two days post invasion. J Nematol 36: 241–248 [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Alkharouf N, MacDonald M, Matthews B. (2005) Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode). Plant Mol Biol 59: 965–979 [DOI] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, Matsye P, Alkharouf NW, Matthews BF. (2009) A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol Biol 71: 525–567 [DOI] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF. (2010) Syncytium gene expression in Glycine max ([PI 88788]) roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol Biochem 48: 176–193 [DOI] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF. (2007a) A time-course comparative microarray analysis of an incompatible and compatible response by Glycine max (soybean) to Heterodera glycines (soybean cyst nematode) infection. Planta 226: 1423–1447 [DOI] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF. (2007b) Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226: 1389–1409 [DOI] [PubMed] [Google Scholar]
- Koenning SR, Wrather JA. (November 2010) Suppression of soybean yield potential in the continental United States from plant diseases from 2006 to 2009. Plant Health Prog http://dx.doi.org/10.1094/PHP-2010-1122-01-RS [Google Scholar]
- Lamb C, Dixon RA. (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lee MW, Jelenska J, Greenberg JT. (2008) Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J 54: 452–465 [DOI] [PubMed] [Google Scholar]
- Lee MW, Lu H, Jung HW, Greenberg JT. (2007) A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol Plant Microbe Interact 20: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Li Y, Chen S, Young ND. (2004) Effect of rhg1 gene on penetration, development, and reproduction of Heterodera glycines race 3. Nematology 6: 727–734 [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Skorupska HT. (1996) Cytological expression of early response to infection by Heterodera glycines Ichinohe in resistant PI 437654 soybean. Genome 39: 986–998 [DOI] [PubMed] [Google Scholar]
- Manickavelu A, Kawaura K, Oishi K, Shin IT, Kohara Y, Yahiaoui N, Keller B, Suzuki A, Yano K, Ogihara Y. (2010) Comparative gene expression analysis of susceptible and resistant near-isogenic lines in common wheat infected by Puccinia triticina. DNA Res 17: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Bastian R, Donaldson L, Murray S, Bajic V, Gehring C. (2008) Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melito S, Heuberger AL, Cook D, Diers BW, MacGuidwin AE, Bent AF. (2010) A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1 locus LRR-kinase on soybean cyst nematode resistance. BMC Plant Biol 10: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL. (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ 4: 671–683 [DOI] [PubMed] [Google Scholar]
- Morse M, Pironcheva G, Gehring C. (2004) AtPNP-A is a systemically mobile natriuretic peptide immunoanalogue with a role in Arabidopsis thaliana cell volume regulation. FEBS Lett 556: 99–103 [DOI] [PubMed] [Google Scholar]
- Mudge J. (1999) Marker assisted selection for soybean cyst nematode resistance and accompanying agronomic traits. PhD thesis. University of Minnesota, St. Paul [Google Scholar]
- Nettleton D. (2006) A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, Shannon JG, Tylka GL. (2002) A revised classification scheme for genetically diverse populations of Heterodera glycines. J Nematol 34: 279–288 [PMC free article] [PubMed] [Google Scholar]
- Niblack TL, Heinz RD, Smith GS, Donald PA. (1993) Distribution, density, and diversity of Heterodera glycines in Missouri. J Nematol 25: 880–886 [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, Innes RW. (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol 144: 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Tachi H, Kojima T, Shiraiwa M, Takahara H. (2006) Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of PR-5 protein in soybean (Glycine max [L.] Merr.). Plant Physiol Biochem 44: 574–580 [DOI] [PubMed] [Google Scholar]
- O’Rourke JA, Nelson RT, Grant D, Schmutz J, Grimwood J, Cannon S, Vance CP, Graham MA, Shoemaker RC. (2009) Integrating microarray analysis and the soybean genome to understand the soybean’s iron deficiency response. BMC Genomics 10: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Potter S, Uknes S, Lawton K, Winter A-M, Chandler D, DiMaio J, Novitzky R, Ward E, Ryals J. (1993) Regulation of a hevein-like gene in Arabidopsis. Mol Plant Microbe Interact 6: 680–685 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Rivas S, Roby D. (2006) An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett 580: 3498–3504 [DOI] [PubMed] [Google Scholar]
- Riggs RD, Kim KS, Gipson I. (1973) Ultrastructural changes in Peking soybeans infected with Heterodera glycines. Phytopathology 63: 76–84 [Google Scholar]
- Ross JP. (1958) Host-parasite relationship of the soybean cyst nematode in resistant soybean roots. Phytopathology 48: 578–579 [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Lin JY, Schlueter SD, Vasylenko-Sanders IF, Deshpande S, Yi J, O’Bleness M, Roe BA, Nelson RT, Scheffler BE, et al. (2007) Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro AD, Zhang C. (2001) The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol 127: 1089–1101 [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade R. (2009) The endoplasmic reticulum stress signaling pathways in plants. Biofactors 35: 326–331 [DOI] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D. (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet A, Hause B, Hause G, Scheel D, Lee J. (2003) The Arabidopsis NHL3 gene encodes a plasma membrane protein and its overexpression correlates with increased resistance to Pseudomonas syringae pv. tomato DC3000. Plant Physiol 132: 2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet A, Parker J, Tornero P, Nass N, Nurnberger T, Dangl JL, Scheel D, Lee J. (2002) NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant Microbe Interact 15: 608–616 [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252 [DOI] [PubMed] [Google Scholar]
- Wang X, Replogle A, Davis EL, Mitchum MG. (2007) The tobacco Cel7 gene promoter is auxin-responsive and locally induced in nematode feeding sites of heterologous plants. Mol Plant Pathol 8: 423–436 [DOI] [PubMed] [Google Scholar]
- Xiao YL, Smith SR, Ishmael N, Redman JC, Kumar N, Monaghan EL, Ayele M, Haas BJ, Wu HC, Town CD. (2005) Analysis of the cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol 139: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Bradshaw JD, Whitham SA, Hill JH. (2010) The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol 153: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang C, Whitham SA, Hill JH. (2009) Development and use of an efficient DNA-based viral gene silencing vector for soybean. Mol Plant Microbe Interact 22: 123–131 [DOI] [PubMed] [Google Scholar]
- Zheng MS, Takahashi H, Miyazaki A, Hamamoto H, Shah J, Yamaguchi I, Kusano T. (2004) Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta 218: 740–750 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]