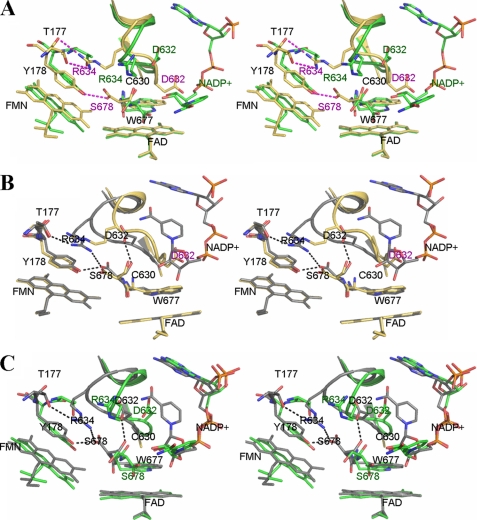
FIGURE 3.
Stereo overlays of the structures of wild type (PDB code 1JA1) (green), 147CC514 (gold), and the binary complex of reduced 147C/C514-NADP+ (gray). For clarity, only the isoalloxazine ring-ribityl portions of flavin cofactors are shown. A, comparison of wild type (green) and 147CC514 (gold with red labels). The NADP+ structure is that found in the wild type structure, because the cross-linked 147CC514 structure lacks bound NADP+. Residues labeled in black are those superimposed in both structures. The loop containing residues Gly631–Asn635 (Asp632 loop) in the structure of 147CC514 is extended down toward where the indole ring of Trp677 lies. This loop movement results in the guanidine group of Arg634 making hydrogen bonds with both the side chain and main chain amide nitrogens of Thr177. Furthermore, the side chain of Asp632 flips down and occupies the space where the ribityl-pyrophosphate moiety of NADP+ would ordinarily bind, causing steric hindrance as well as charge repulsion between the NADP+ pyrophosphate and Asp632 carboxylate, preventing NADP+ from binding to 147CC514. B, overlay of the structure of NADP+-free 147CC514 (gold) and that of NADP+ bound disulfide-reduced 147CC514 (gray). Upon binding of NADP+, the Asp632 loop of 147C/C514 is pushed further toward the FMN domain by as much as 6 Å for the Ala633 Cα atom and 4 Å for the Asp632 Cα, making room for NADP+ to bind. Arg634 makes a salt bridge with the C-terminal carboxylate of Ser678. As shown in A, Asp632 of 147CC514, which is located in the middle of the Asp632 loop, may impair NADP+ binding by both steric hindrance and charge repulsion. C, comparison between wild type (green) and 147C/C514-NADP+ (gray). Both the Asp632 loop and NADP+ conformations are different between these two structures; an extended NADP+ conformation is found in the wild type structure (green carbons with red oxygen and phosphorus atoms), although a more compact, folded conformation is observed in the mutant structure (gray carbons).