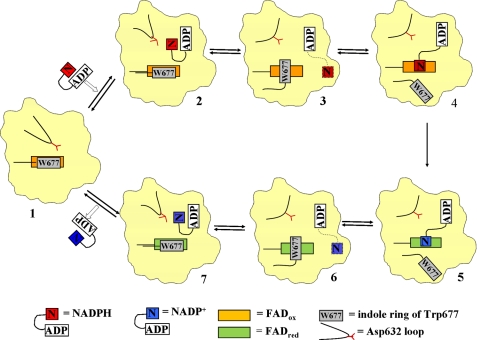
FIGURE 8.
Schematic illustration of NADPH binding to and NADP+ release from CYPOR. For clarity, only the FAD/NADP(H)-binding domain of CYPOR is shown. NADPH enters oxidized CYPOR (state 1, modeled after the structure of 147CC514, which lacks bound NADP(H)). NADPH initially binds to the enzyme in a folded conformation, such that the adenine ring and the nicotinamide ring almost stack on each other. Upon binding of NADPH, the Asp632 loop of CYPOR moves away to allow NADPH to bind (state 2, modeled from the structure of 147C/C514-NADP+). Once the NADPH binds, the Asp632 loop moves in and displaces the nicotinamide ring, resulting in the ribityl-nicotinamide moiety extending to search for the proper binding site for hydride transfer, while keeping the ADP-PPi anchor at the arginine site. The indole ring of Trp677 rotates to be ready to move away from the flavin ring (state 3, the structure of wild type; PDB code 1JA1). The indole ring moves away to make room for the nicotinamide ring to bind at the re-side of the isoalloxazine ring (state 4, modeled after the structure of the mutant W677X, in which the two penultimate residues of CYPOR, Trp677 and Ser678, were truncated, PDB code 1JA0). Hydride transfer occurs; FAD is reduced, and NADPH becomes NADP+ (state 5, from PDB code 1JA0). Once the flavin is reduced and the nicotinamide is oxidized, the nicotinamide ring moves out and the indole ring returns to the re-face of the FAD ring (state 6, PDB code 1JA1). As the Asp632 loop moves away allowing for the nicotinamide ring to fold back, NADP+ adopts the folded conformation poised to dissociate from the enzyme (state 7, the structure of 147C/C514-NADP+). The Asp632 loop moves back closer to where the NADP+ pyrophosphate-2′-AMP lies, causing steric hindrance as well as electrostatic repulsion, resulting in dissociation of the cofactor from CYPOR. The enzyme now returns to state 1 and the cycle repeats.