Abstract
Background
The human pathogen Listeria monocytogenes resides and proliferates within the cytoplasm of epithelial cells. While the virulence factors essentially contributing to this step of the infection cycle are well characterized, the set of listerial genes contributing to intracellular replication remains to be defined on a genome-wide level.
Results
A comprehensive library of L. monocytogenes strain EGD knockout mutants was constructed upon insertion-duplication mutagenesis, and 1491 mutants were tested for their phenotypes in rich medium and in a Caco-2 cell culture assay. Following sequencing of the plasmid insertion site, 141 different genes required for invasion of and replication in Caco-2 cells were identified. Ten in-frame deletion mutants were constructed that confirmed the data. The genes with known functions are mainly involved in cellular processes including transport, in the intermediary metabolism of sugars, nucleotides and lipids, and in information pathways such as regulatory functions. No function could be ascribed to 18 genes, and a counterpart of eight genes is missing in the apathogenic species L. innocua. Mice infection studies revealed the in vivo requirement of IspE (Lmo0190) involved in mevalonate synthesis, and of the novel ABC transporter Lmo0135-0137 associated with cysteine transport. Based on the data of this genome-scale screening, an extreme pathway and elementary mode analysis was applied that demonstrates the critical role of glycerol and purine metabolism, of fucose utilization, and of the synthesis of glutathione, aspartate semialdehyde, serine and branched chain amino acids during intracellular replication of L. monocytogenes.
Conclusion
The combination of a genetic screening and a modelling approach revealed that a series of transporters help L. monocytogenes to overcome a putative lack of nutrients within cells, and that a high metabolic flexibility contributes to the intracellular replication of this pathogen.
Background
Listeria monocytogenes is a gram-positive, food-borne pathogen which is able to grow at low temperatures down to 1.7°C [1]. It is widely distributed in nature and mainly affects immunocompromised individuals. A hallmark of this facultative intracellular pathogen is its capability to use eukaryotic cells as a predominant growth niche [2].
L. monocytogenes entry into epithelial cells is mediated by several cell surface proteins including internalin A and B [3]. The phagosomal membrane is then disrupted by the activity of a hemolysin and phospholipases, resulting in the release of L. monocytogenes into the cytosol of its host cell where it encounters an environment of undefined composition. There, the pathogen is able to move intra- and intercellular by expressing ActA which polymerizes host actin molecules. The genes required for these steps of the infection cycle are under control of the central transcription regulator PrfA [4]. While these virulence factors have been extensively studied in the last years, far less is known about the availability of nutrients in the cytosol of eukaryotic cells, and the specific metabolic adaptations of L. monocytogenes including substrate acquisition that allow its efficient proliferation within this cellular compartment [2,5,6]. The knowledge of the gene set involved in the multiplication of L. monocytogenes inside epithelial cells might also help to discover its metabolic Achilles' heel as a prerequisite to control its intracellular lifestyle and to combat this medically important pathogen.
In a pilot study on virulent strains or mutants of L. monocytogenes auxotrophic for amino acids and nucleobases, it was revealed that hosts can provide sufficient organic and inorganic compounds to overcome selected auxotrophies with respect to uracil, phenylalanine, glycine, proline, or nicotinic acid [7]. In contrast, the synthesis of all three aromatic amino acids and adenine is essential for efficient cytosolic replication in J774A.1 macrophage cells. Some further factors involved in nutrient uptake and metabolism have also been shown to be necessary for intracellular survival and proliferation of pathogenic L. monocytogenes. By screening a library of Tn917-lac insertion mutants, the specific induction of genes for purine and pyrimidine biosynthesis as well as for arginine uptake in macrophages was observed [8]. Reduced replication has been demonstrated for a mutant of the PrfA-dependent gene hpt whose product is involved in the exploitation of hexose phosphate from the host cell [9]. The use of host-derived lipoic acid has also been shown to be required for intracellular replication and virulence of L. monocytogenes. A mutant lacking the lipoate protein ligase LplA1 was less virulent in the mouse model, probably due to a loss of pyruvate dehydrogenase function whose E2 subunit is modified by LplA1 [10]. In two independent transcriptomic approaches following infection of macrophages and epithelial cells, it was demonstrated that the pentose phosphate cycle, but not glycolysis is the predominant pathway of listerial metabolism in the cellular host environment [11,12]. Induced synthesis of the branched chain amino acids and the utilization of alternative carbon sources such as glycerol were also observed in these studies. The absence of the Entner-Doudoroff pathway and a bifurcated citric acid cycle due to the lack of α-ketoglutarate dehydrogenase activity were predicted by genome analysis and then experimentally demonstrated [13,14]. Little is known about the in vivo nitrogen metabolism of L. monocytogenes that is assumed to use ammonium, arginine or ethanolamine as an alternative nitrogen source during intracellular replication [12]. Results with amino acids-deprived tissue culture cells and a L. monocytogenes mutant auxotrophic for threonine indicate that this pathogen may utilize intracellular peptides as a source of amino acids [7]. This assumption is supported by oppA encoding an oligopeptide permease that is involved in intra-macrophagic survival, indicating that the efficient uptake of oligopeptides favours growth of L. monocytogenes in macrophages [15]. Listerial growth within eukaryotic cells also depends on vitamins. Both the uptake of thiamine and the de novo biosynthesis of its precursors have recently been shown to contribute to intracellular proliferation, a finding that reflects the role of thiamine as a cofactor of enzymes involved in central metabolic processes such as the pentose-phosphate cycle or the synthesis of branched chain amino acids [16]. Furthermore, the listerial genome harbours the genes for cobalamine biosynthesis required for intracellular ethanolamine degradation [12,17].
To obtain an overview of genes required by L. monocytogenes to efficiently replicate in mammalian cells, we established a mutant library of L. monocytogenes by insertion duplication mutagenesis (IDM) and screened 1491 mutants for impaired capacity to invade or replicate within Caco-2 cells. A total of 141 mutants were isolated, most of them involved in metabolism, in transport and in cell wall functions. All mutants showed wildtype-like growth in nutrient-rich medium, indicating the compartment-specific role of the genes tested. Mathematical modelling then allowed to improve our understanding of the complex data obtained. The overall results shed further light on the set of listerial genes required for the replication in the cytosol of epithelial cells, and on the environment that L. monocytogenes encounters within host cells.
Results
141 genes were identified that affect cell invasion or intracellular replication
For the identification of insertional mutations that affect the ability of L. monocytogenes EGD to proliferate within eukaryotic cells, 1491 L. monocytogenes insertion mutants were screened for a phenotype in a Caco-2 cell infection assay. 238 mutants that exhibited invasion defects or reduced intracellular replication rates in at least three independent experiments were identified. As the insertion of pLSV101 into the chromosome leads to slight growth deficiencies, the intracellular growth data of a random group of more than 100 IDM mutants were collected [12], and an at least two-fold attenuation with respect to the average data of the control group was set as a threshold. All mutants were then grown in BHI medium to exclude unspecific growth deficiencies, and only mutants exhibiting wildtype-like growth were considered for further analysis. Sequence analysis revealed that in these mutants, 141 different gene loci had been targeted by IDM (Table 1). The remaining mutants carried insertions of identical fragments or fragments of genes that had already been identified. As indicated in Table 1, 23 of the genes identified have recently been described due to the fact that their transcription is upregulated during replication of L. monocytogenes in Caco-2 cells or macrophages, while the expression of 19 other genes is reduced in these eukaryotic cells [11,12].
Table 1.
Genes identified to be required for intracellular replication of L. monocytogenes within epithelial cells
| fold reduction | fold reduction | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| gene name or number | protein name, or protein homology/similarity to | polar effect | intrac. | invasion/adhesion | gene name or number | protein name, or protein homology/similarity to | polar effect | intrac. | invasion/adhesion |
| cell wall | protein modification | ||||||||
| 1lmo0441 | penicillin-binding protein (D-alanyl-D-alanine carboxypeptidase) | no | 9.4 | 6.1 | 5lmo0618 | protein kinase | no | 11.0 | 2.0 |
| 2,8lmo1085 | teichoic acid biosynthesis protein B | rf | 12.3 | 2 | 1lmo0763 | hypothetical Ser/Thr protein phosphatase family protein | possible | 5.5 | 2.0/5.0 |
| lmo1088 | teichoic acid biosynthesis protein B | rf | 26.2 | 1.9 | metabolism of amino acids and related molecules | ||||
| 7lmo1713 | actin-like ATPase involved in cell morphogenesis | no | 3.6 | 3.5 | lmo0594 | homoserine O-acetyltransferase | no | 7.7 | 3.9 |
| 3,5,7pbpB (lmo2039) | penicillin-binding protein 2B | rf | 3.8 | 2.6 | 6,8lmo1235 | aspartokinase II a subunit | no | 5.9 | nd |
| 1lmo2555 | Glycosyltransferase | rf | 8.1 | 3.9/5.0 | lmo1495 | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | possible | 6.2 | 1.0 |
| transport/binding proteins and lipoproteins | lmo1916 | peptidase | no | 7.0 | 2.4 | ||||
| 4lmo0135 (ctpA) | oligopeptide ABC transport system, substrate binding protein | rf | 13.6 | 2.2 | 8aroB (lmo1927) | 3-dehydroquinate synthase | rf | 2.8 | 1.4 |
| 4lmo0136 | oligopeptide ABC transport system, substrate binding protein | rf | 5.0 | 2.4 | 4,6ilvB (lmo1484) | acetolactate synthase (acetohydroxy-acid synthase) | rf | 5.5 | 1.8 |
| lmo0137 | oligopeptide ABC transport system, permease | no | 9.7 | 1.9 | 4,6ilvC (lmo1486) | ketol-acid reductoisomerase | no | 3.2 | 3 |
| 4lmo0195 | ABC-type antimicrobial peptide transport system, permease | no | 6.3 | nd | lmo2051 | weakly similar to proteases | no | 4.1 | 1 |
| lmo0495 | permease of the drug/metabolite transporter (DMT) superfamily | possible | 4.2 | 2.3 | lmo2694 | lysine decarboxylase | possible | 5.5 | 0 |
| 4lmo0584 | conserved hypothetical membrane protein, putative permease | no | 7.2 | 3.0 | lmo2770 | γ-glutamylcysteine synthetase and cyanophycin synthetase | no | 5.8 | 2.3 |
| lmo0645 | amino acid transporter | no | 4.9 | 1.5 | 6serC (lmo2825) | phosphoserine aminotransferase | possible | 5.7 | 3.5 |
| 5lmo0650 | conserved membrane protein | possible | 2.8 | 2.0 | metabolism of nucleotides and nucleic acids | ||||
| lmo0787 | amino acid transporter | no | 3.4 | 1.0 | 1,5purA (lmo0055) | adenylosuccinate synthetase | no | 14.9 | /4.0 |
| lmo0810 | spermidine/putrescine-binding protein | no | 7.4 | 3.1 | purQ (lmo1769) | phosphoribosylformylglycinamidine synthetase | rf | 12.0 | nd |
| 3lmo1003 | phosphotransferase system enzyme I | no | 3.4 | 1.0 | 8purS (lmo1771) | phosphoribosylformylglycinamidine synthetase | rf | 11.0 | 1.0 |
| gbuA (lmo1014) | glycine betaine ABC transporter, ATP-binding protein | rf | 10.0 | 4.0 | pyrE (lmo1831) | orotatephosphoribosyltransferase | no | 5.5 | 2.5 |
| 8lmo1416 | hypothetical transporter | no | 5.3 | 1.0 | metabolism of lipids | ||||
| 6opuCA (lmo1428) | glycine betaine/carnitine/choline ABC transporter, ATP-binding protein | rf | 15.7 | 1.7 | 5,8ispE (lmo0190) | CDP-ME synthase involved in isoprenoid biosynthesis | no | 4.7 | 2.3 |
| 5lmo1431 | ABC transporter, ATP binding protein | no | 4.8 | 3 | lmo1005 | 3-hydroxyisobutyrate dehydrogenase | no | 11.0 | 1.8 |
| lmo1506 | ABC-type antimicrobial peptide transport system, permease | rf | 6.2 | 2.3 | lmo1363 | geranyltransferase | no | 5.0 | 2.8 |
| lmo1739 | amino acid ABC transporter | rf | 5.5 | 2 | lmo2450 | carboxylesterase | possible | 4.2 | 3.0 |
| 3,5,7,8lmo1847 | ABC transporter specific for metal cations | no | 5.8 | 2.3 | metabolism of coenzymes and prosthetic groups | ||||
| lmo2124 | maltodextrin ABC transport system, permease | rf | 3.4 | 0 | 5lmo0221 | hypothetical type III pantothenate kinase | possible | 6.9 | 1.0 |
| 1,3oppF (lmo2192) | oligopeptide ABC-transporter, ATP-binding protein | no | 21.0 | 8.0/4.0 | 1,3pdxK (lmo0662) | pyridoxine kinase | no | 13.1 | 1/4.0 |
| lmo2227 | ABC transporter, ATP-binding protein | possible | 5.8 | 1.3 | lmo1043 | molybdopterin-guanine dinucleotide biosynthesis MobB | rf | 7.9 | 1.4 |
| lmo2249 | low-affinity inorganic phosphate transporter | no | 2.8 | 2.0 | lmo1932 | heptaprenyl diphosphate synthase component I | rf | 12.1 | 4.8 |
| lmo2353 | hypothetical Na+/H+ antiporter | no | 4.3 | 2.3 | nadB (lmo2023) | L-aspartate oxidase | rf | 3.4 | 0 |
| lmo2380 | protein involved in resistance to cholate/Na+ and in pH homeostasis | rf | 5.7 | 2.5 | 3lmo2102 | glutamine amidotransferase subunit PdxT (pyridoxine biosynthesis) | no | 4.0 | 3.3 |
| lmo2430 | B. subtilis ferrichrome ABC transporter FhuG, permease | rf | 75.6 | 5.0 | lmo2566 | biotin/lipoate A/B protein ligase family | possible | 6.2 | 2.0 |
| lmo2816 | transport protein | no | 6.3 | 2.0 | DNA restriction/modification and repair | ||||
| lmo2850 | sugar transport protein | rf | 16.5 | 0 | 5gyrB (lmo0006) | DNA gyrase subunit B | rf | 9.6 | nd |
| sensors | lmo0157 | ATP-dependent helicase | possible | 2.1 | 1.0 | ||||
| lmo0799 | oxygen/light sensor with PAS domain | no | 8.3 | 2.0 | mfd (lmo0214) | transcription-repair coupling factor | possible | 8.9 | 3.7 |
| lmo1508 | two-component sensor histidine kinase | no | 5.2 | 2.9 | lmo0588 | DNA photolyase | no | 9.6 | 2.0 |
| membrane bioenergetics | mutM (lmo1564) | formamidopyrimidine-DNA glycosylase | no | 5.2 | 1.1 | ||||
| lmo0091 | ATP synthase g chain, H+-transporting two-sector ATPase | rf | 5.7 | 2.0 | lmo1751 | hypothetical RNA methyltransferase | no | 6.2 | 2.0 |
| nifJ (lmo0829) | pyruvate-flavodoxin oxidoreductase | no | 6.4 | 3.6 | 8ansB (lmo1663) | asparaginyl-tRNA synthetases | no | 4.1 | 1.6 |
| 5atpA (lmo2531) | H+-transporting ATP synthase chain α | rf | 7.7 | 2.7 | lmo2050 | exconuclease ABC (subunit A) | no | 5.1 | 2.0 |
| atpB (lmo2535) | H+-transporting ATP synthase chain β | rf | 4.4 | 5.9 | DNA recombination | ||||
| mobility and chemotaxis | 6recN (lmo1368) | RecN | no | 14.8 | 1.9 | ||||
| lmo0680 | flagella-associated protein FlhA | rf | 7.0 | 3.1 | ruvB (lmo1532) | Holliday junction DNA helicase RuvB | possible | 9.4 | 3.9 |
| 6lmo0700 | flagellar motor switch protein FliY | rf | 5.8 | 1.7 | 8recS (lmo1942) | similar to ATP-dependent DNA helicase | no | 7.0 | 2.3 |
| cell surface proteins | regulation | ||||||||
| 1,2,6vip (lmo0320) | putative peptidoglycan bound protein with LPXTG motif | no | 6.4 | 4.5 | 3agrA (lmo0051) | 2-component response regulator protein | no | 3.4 | 1.0 |
| 8lmo0327 | protein with LPXTG motif, putative murein hydrolase activity | no | 6.9 | 3.0 | 6lmo0294 | transcription regulator, LysR-gltR family | no | 7.4 | 3.1 |
| 4lmo0514 | internalin-like protein with LPXTG motif | no | 6.4 | 1.4 | lmo0535 | transcription regulator, LacI family | no | 9.5 | 1.0 |
| 4lmo0576 | hypothetical cell wall associated protein | no | 4.2 | 1.5 | 3fur (lmo1956) | transcriptional regulator, Fur family | no | 5.5 | 1.0 |
| 2,8lmo1666 | peptidoglycan linked protein with LPXTG motif | no | 6.8 | 1.8 | lmo1994 | transcription regulators, LacI family | no | 9.0 | 1.4 |
| lmo2026 | hypothetical peptidoglycan bound protein with LPXTG motif | no | 8.7 | 1.4 | RNA modification | ||||
| 5lmo2504 | cell wall binding protein, peptidase-related enzyme | no | 4.8 | 2.0 | lmo0241 | hypothetical RNA methyltransferase, trmH family protein | rf | 4.1 | 1.0 |
| 1ami (lmo2558) | autolysin, N-acetylmuramoyl-L-alanine amidase | no | 3.8 | 5.3 | 1,3,5lmo1434 | RNA-metabolising metallo-β-lactamase | no | 9.6 | 3/22 |
| metabolism of carbohydrates and related molecules | miscellaneous | ||||||||
| glpQ (lmo0052) | transmembrane protein with phosphoesterase domain | possible | 4.1 | 2.0 | lmo0066 | toxin component of A/B toxin | rf | 2.8 | 1.0 |
| 8lmo0182 | α-xylosidase and α-glucosidase | rf | 7.9 | 1.3 | 4lmo0585 | secreted protein | no | 4.8 | 1.0 |
| lmo0261 | phospho-β-glucosidase | no | 3.4 | 0 | lmo0587 | secreted protein, YapH from Y. pestis, cell wall surface protein | no | 3.4 | 1.0 |
| 8lmo0271 | phospho-β-glucosidase | possible | 4.1 | 1.0 | unknown proteins | ||||
| lmo0517 | phosphoglycerate mutase | no | 13.8 | 2.3 | lmo0276 | hypothetical hydrolase, HAD superfamily | possible | 5.2 | 1.3 |
| 2lmo1031 | hypothetical L-fucose isomerase | rf | 11.1 | 2.1 | 2lmo0313 | hypothetical hydrolase, PHP superfamily | no | 2.8 | 1.0 |
| 2lmo1032 | Transketolase | rf | 14.4 | 4lmo0590 | hypothetical DAK2/DegV domain-containing protein | possible | 9.6 | 1.0 | |
| lmo1166 | NADPH-dependent butanol dehydrogenaseI | possible | 4.2 | 1.0 | lmo0765 | unknown protein | possible | 10.4 | 3.0 |
| 4glpD (lmo1293) | glycerol-3-phosphate dehydrogenase | no | 4.8 | 1.0 | 4,6,7lmo0794 | B. subtilis YwnB protein | no | 12.4 | 2.0 |
| 6lmo1244 | weakly similar to phosphoglycerate mutase 1 | no | 8.3 | 2.0 | lmo1379 | B. subtilis SpoIIIJ protein, hypothetical membrane protein | no | 4.8 | 3.0 |
| lmo2005 | Oxidoreductase | no | 7.4 | 3.9 | 3lmo1402 | B. subtilis YmcA protein | no | 2.1 | 2.0 |
| lmo2015 | α-mannosidase | no | 6.4 | 1.7 | lmo1575 | phosphoesterase, DHH superfamily | no | n.d. | 1.0 |
| lmo2134 | fructose-1,6-biphosphate aldolase type II | no | 5.2 | 2.4 | 6,8lmo1700 | unknown protein | no | 4.6 | 1.3 |
| lmo2172 | propionate CoA-transferase | possible | 5.3 | 1.3 | 4lmo1830 | short chain dehydrogenase | no | 11.7 | 2.0 |
| lmo2247 | Oxidoreductase | possible | 5.0 | 2.4 | lmo1866 | hypothetical phosphotransferase | possible | 7.1 | 2.2 |
| 8lmo2446 | Glycosidase | no | 9.9 | 2.5 | lmo1920 | unknown protein | 16.5 | 1.0 | |
| lmo2586 | formate dehydrogenase α-chain | possible | 2.4 | 2.2 | lmo2516 | conserved hypothetical protein | possible | 3.6 | 1.2 |
| 6lmo2660 | Transketolase | rf | 3.0 | 2.6 | lmo2639 | unknown protein, contains DUF1312 domain | possible | 5.5 | 1.0 |
| lmo2764 | xylose operon regulatory protein and to glucose kinase | rf | 3.6 | 2.1 | no similarity | ||||
| 2lmo2781 | β-glucosidase | rf | 6.5 | 2.0 | lmo0729 | no similarity | no | 4.8 | 1.0 |
| lmo2831 | Phosphoglucomutase | no | 5.3 | 2.8 | 2lmo1188 | no similarity | no | 5.1 | 3.5 |
| lmo1219 | no similarity | no | 4.8 | 2.0 | |||||
| lmo2129 | no similarity | no | 5.8 | 1.6 | |||||
1adhesion (seven genes) or invasion (four genes) defective as determined 35 min or 2 h post infection, respectively; 2genes without homologue in L. innocua; genes downregulated3 or upregulated4 during growth in macrophages [11]; genes downregulated5 or upregulated6 during growth in Caco-2 cells [12]; 7induced in vivo [23]; 8prfA binding box is located upstream [13]. Fold reduction of intracellular (intrac.) replication in comparison to a control group of >100 insertion mutants [12]. rf, related function of downstream genes. Four genes from categories which one gene only (1.6 protein secretion, 1.7 cell division, 1.10 transformation, and 3.7 protein synthesis) was assigned to are not shown. See also Additional file 6 for generation times.
Validation by in-frame deletions
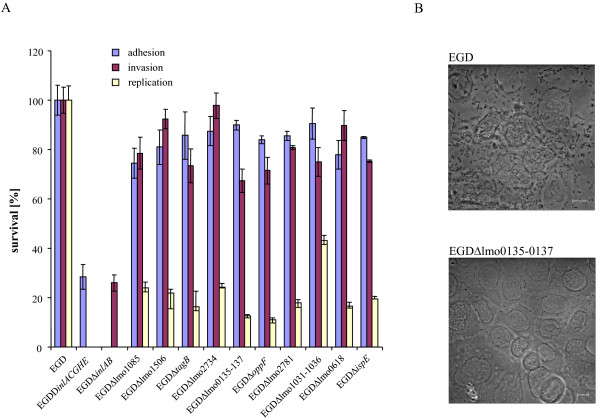
Although IDM has the potential for termination-induced reduction of downstream gene expression [18], a real-time RT-PCR approach revealed that regardless of its orientation, plasmid pLSV101 insertion did not significantly affect the transcription of the most distantly located genes of the operons investigated [12]. Moreover, genes clustered in an operon are often involved in the same cellular pathway or function (Table 1). To nevertheless further validate the data obtained, a series of ten non-polar deletion mutants were constructed. With the exception of lmo0618 expressing a putative protein kinase, genes selected for deletion encode proteins that belong to the classes of cell wall biosynthesis, metabolism, and transport. In all but one case, a five- to tenfold reduction in levels of intracellular replication within Caco-2 cells in comparison to the wildtype strain was observed (Figure 1A). EGDΔlmo1031-1036 showed a more modest reduction of replication, thus confirming the result of a recent study on listerial glycerol metabolism [19]. EGDΔlmo0135-0137 is an example for a very strong attenuation of the intracellular proliferation rate (Figure 1B). The experiments were also performed with an MOI of approximately 100, resulting in similar intracellular attenuation rates (data not shown). Taken together, each of the in-frame deletions led to intracellular growth attenuation, thus confirming the mutagenesis strategy applied.
Figure 1.
Intracellular replication of L. monocytogenes deletion mutants. (A) Caco-2 cells were infected at a multiplicity of infection (MOI) with either the wildtype strain or in-frame deletion mutants. The mutants EGDΔinlACGHE and EGDΔinlAB served as controls for the adhesion and invasion properties of the mutants. The number of viable bacterial cells recovered from the epithelial cells 8 h after infection was determined, and reduced survival of the ten mutants was calculated as a percentage in comparison with the wildtype strain. Error bars show the standard deviations from the mean. Each experiment was performed independently at least three times. The significance level was < 0.05 according to student's t test. (B) Phase contrast microscopy of a Caco-2 cell layer 8 h after infection with the wildtype strain (top) and mutant EGDΔlmo0135-0137 (buttom). The scale bars corresponds to 10 μm. The deletion of the transporter operon results in an approximately 10-fold reduced number of L. monocytogenes cells.
Classification of listerial genes required for invasion of and replication in Caco-2 cells
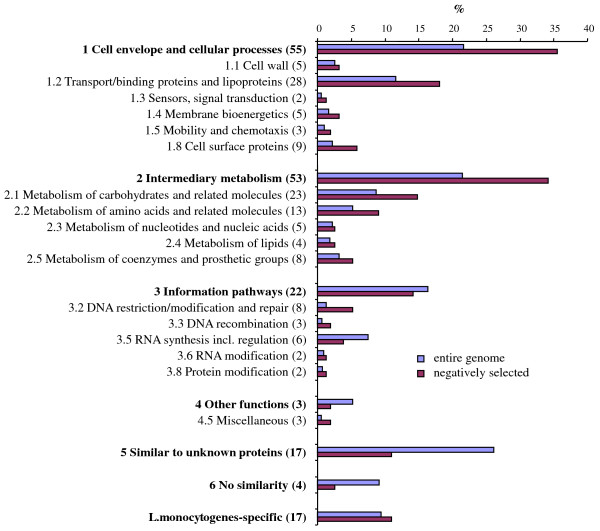
All genes were classified into functional categories (Figure 2, Table 1). 18 insertions into functional unknown genes were identified. Another striking feature of the classification is that 27 of the mutated genes belong to the class of transporters and lipoproteins, among them several uptake systems for sugars. At least five of the transporters isolated are possibly involved in the uptake of amino acids and oligopeptides (Table 1). Two of a total of ten antimicrobial permeases encoded by the listerial genome were also identified. A huge set of 47 genes are involved in intermediary metabolism, among them 21 genes contributing to the utilization of carbohydrates such as glucose, glycerol, or fucose, and 11 to the metabolism of amino acids (Table 1). Interestingly, genes of the two groups cell envelope/cellular processes including transport (35%) and intermediary metabolism (34%) are overrepresented, and genes of unknown function (11%) underrepresented with respect to their quota in the entire genome (22%, 22%, and 26%, Figure 2).
Figure 2.
Distribution of 157 genes negatively selected within different functional categories. Classification and categorization of genes was done essentially in accordance with Glaser et al. [13] and the L. monocytogenes genome sequence provided by the Pasteur Institute http://genolist.pasteur.fr/ListiList/. Categories which no gene or one gene only (1.6 protein secretion, 1.7 cell division, 1.10 transformation, 3.7 protein synthesis) was assigned to were not listed, but the genes are included in total numbers (bold letters). For each category, the percentage of gene numbers relative to the total gene number in the L. monocytogenes genome (blue) and the number of genes with a Caco-2 infection phenotype relative to the total number of genes selected (red) is shown. For each class and category, the number of genes identified is indicated in brackets. 16 genes only recently identified [12] are included.
Eight genes identified in the screening described here have no homologue in the apathogenic species L. innocua (Table 1), indicating that their products might specifically contribute to virulence properties of L. monocytogenes. This group includes one L. monocytogenes-specific gene (lmo1188), lmo1085 required for teichoic acid synthesis, and several metabolic genes such as lmo2781 encoding a glucosidase (see below) or lmo1031 possibly involved in fucose utilization.
Genes involved in invasion of epithelial cells, and cell surface genes
To discriminate between factors playing a role during intracellular multiplication from those that contribute to Caco-2 cell adhesion or invasion, the number of viable surface-attached or intracellular L. monocytogenes cells 35 minutes or two hours after infection was determined for all mutants selected above. Only mutants affected in their adhesion or invasion capabilities are expected to be identified by this approach. As bacterial entrance is a continuous process and cannot strictly be separated from the intracellular replication phase, only four mutants with an at least five-fold reduced number of intracellular bacteria two hours post infection were classified as invasion deficient. Further analysis of the plasmid insertion site revealed ami, lmo0441 encoding a putative D-alanyl-D-alanine carboxypeptidase, oppF encoding a membrane protein with similarity to an ABC transporter, and vip (lmo0320) (Table 1).
Modelling
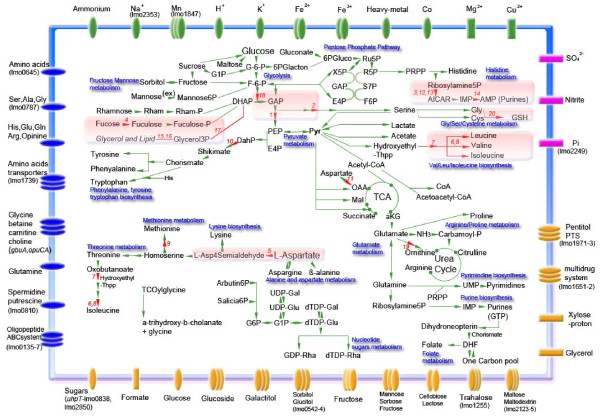
The main anabolic and catabolic pathways of L. monocytogenes such as glycolysis, starch degradation, the pentose phosphate pathway, the tricarboxylic acid cycle (TCA), the metabolism of lipids and nucleic acids, and the biosynthesis of essential amino acids were reconstructed as described above, including consideration of important transporters (Figure 3). Upon data mining, additional genes were found in the literature to play a role during intracellular replication. 31 of them were further considered due to the availability of quantitative data (see Additional file 1 for list of genes and references). From Table 1 and Additional file 1, 20 genes where selected whose mutation resulted in an at least 3.2-fold reduction of the intracellular replication rate (see Additional file 2 for list of genes) and used for a simulation of the listerial metabolism during intracellular replication (see Additional file 3 for input data of the model for calculation with flux balance analysis).
Figure 3.
Modelling of the listerial metabolism during intracellular replication. The main anabolic and catabolic pathways of L. monocytogenes are shown. Transporters for cations (green), anions (pink), carbohydrates and organic acids (orange), and amino acids and peptides (blue) are depicted in the membrane. Numbers in red indicate key affected enzymes, areas shaded in light red key pathways for intracellular survival according to this study. Transporters are generally referred to TransportDB [56] and the Transport Classification Database tcdb [57], their known specificities were discussed [58]. Note the incomplete TCA pathway and a short-cut in the urea cycle [13,14]. Abbreviations: AICAR, 5-amino-1-(5-phospho-D-ribosyl)-imidazole-4-carboxamide; E4P, erythrose-4-phosphate; DHF, dihydrofolate; G-1-P, glucose-1-phosphate; DahP, 3-deoxy-D-arabino-heptulosonate-7-phosphate; F-6-P, fructose-6-phosphate; G-6-P, glucose-6-phosphate; GAP, glyceraldehyde-3-phosphate; OAA, oxaloacetate; aKB, 2-oxoglutarate; 6PGluco, 6-phosphogluconate; PRPP, 5-phosphoribosyl 1-pyrophosphate; R5P, ribose-5-phosphate; Ru5P, ribulose-5-phosphate; S7P, sedaheptulose-7-phosphate; TCA, tricarboxylic acid; TCOylcine, 3α,7α,12α-trihydroxy-5β-cholan-24-oylglycine; X5P, xylulose-5-phosphate; L-Asp4Semialdehyde, L-Aspartate 4-semialdehyde; GSH, glutathione.
Flux balance analysis [20] was applied to identify pathways critical for intracellular survival. Flux balance analysis assumes a steady state for all metabolites internal to the system. Given all biochemical reactions in a network, for each flux pathway in the network an equilibrium condition for each internal metabolite used by the different enzymes of this pathway has to be satisfied. Solving this balancing condition for all biochemical reactions in the network, the so called stochiometric matrix, directly enumerates all possible stable pathways of the system allowing equilibrium for their respective internal metabolites. Two types of calculation are possible: (i) All possible pathways in the system which satisfy the balancing condition and cannot be split further. These are used for elementary mode analysis (EMA). (ii) The extreme pathway analysis (EPA): a minimal set of pathways (a subset derived from EMA) that can completely describe the whole system by linear combinations of these fewer EPA pathways. They are termed extreme pathways because unique use of one of these pathways marks a boundary or extreme situation of the complete space of possibilities. While EPA is faster, one looses some solutions from EMA, in particular all those elementary pathways that are not at the boundary of the system but within it.
The metabolic model allows to identify all cellular pathways affected by each knockout mutation. Applying PERL scripts, we identified the number of mutants impairing the same pathway. Finally, all pathways relevant to intracellular survival were ranked with the highest redundancy at the top, and the key enzymes involved were listed (see Additional file 4 for results obtained from the knockout in silico experiment). A condensed view of these results is given in Figure 3.
Metabolic pathways severely affected by knockout mutations are the biosynthesis of valine/leucine/isoleucine, the purine, fucose, glycerol and lipid metabolism, lower glycolysis as well as serine and glutathione production and aspartate semialdehyde biosynthesis. It is important to note that on the other hand, a huge landscape of central metabolism is not important for intracellular growth. This demonstrates not only the robustness of these central metabolic pathways, but also the difference in the phenotypes revealed by the comparison of growth in full medium with the intracellular replication. The overview from EPA indicates that the glycerol metabolism is most critical for intracellular survival. Further genes important for intracellular replication of L. monocytogenes are serC, lmo0517, glpD and three genes involved in purine synthesis (purS, purQ and purE). The gene lmo0517 encodes a critical enzyme for glycolysis (periplasmic phosphoglycerate mutase), glpD encodes a glycerol-3-phosphate dehydrogenase which plays an important role in the lipid metabolism. In addition to serC, the purine operon, and lmo0517 (pgm), the more extensive EMA also suggests aroB to be critical for the intracellular survival. In addition to the result of EPA, glpD revealed to be less important for the system. This discrepancy might be explained by considering glycerol-3-phosphate as an intermediate metabolite for the glycolysis. On the other hand, lmo1031 putatively involved in fucose metabolism appears more important than argD for the cytoplasmic survival of L. monocytogenes as determined by EMA.
IspE and the transporter Lmo0135-0137 are required in vivo
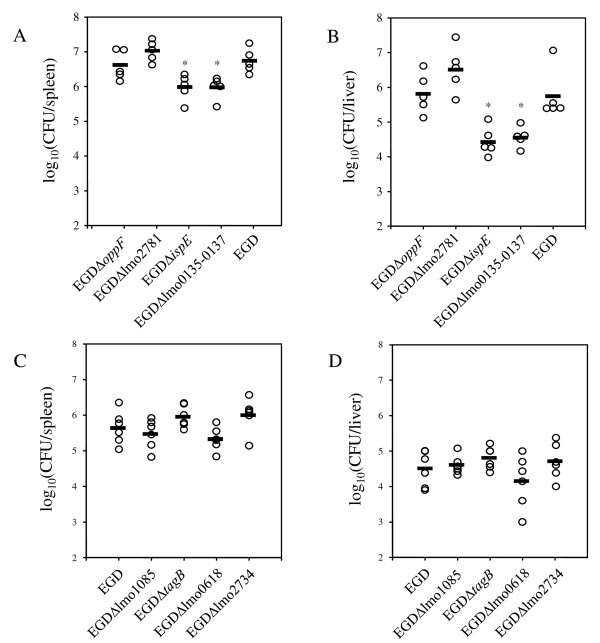
L. monocytogenes mutants exhibiting intracellular growth deficiencies are often attenuated in vivo. Eight of the deletion mutants described above were therefore tested for their virulence properties in the BALB/c mouse infection model. For this purpose, groups of five 8- to 10-week-old female BALB/c mice were infected as described with a sub-lethal dose of the deletion mutants. Mice generally showed some signs of disease after three days of infection. At day three post infection with EGDΔlmo0135-0137 and EGDΔispE, the bacterial load of mice was significantly (P < 0.05) reduced in spleen and liver compared to mice infected with the wildtype-strain (Figure 4). The reduction was approximately one log10 in the spleen (Figure 4A) and more than 1.5 log10 in the liver (Figure 4B), respectively. We also determined the bacterial loads of mice infected with the strains EGDΔlmo0135-0137 and EGDΔispE six days after infection, and again observed an attenuation of these mutants compared to the wildtype strain (data not shown). Gene ispE (lmo0190) encodes a 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate (CDP-ME) synthase involved in the alternative non-mevalonate (MEP) pathway of isoprenoid biosynthesis [21]. The operon lmo0135-0137 encodes a cysteine uptake-associated ABC-transporter [22]. It corresponds to a number of modes in the model (see Additional file 3 for input data of the model for calculation with flux balance analysis), thus supporting the importance of this route for the metabolic network.
Figure 4.
Virulence of L. monocytogenes mutants in vivo. The bacterial loads of infected mice were determined three days and six days after intravenous infection with a sublethal dose of 2 × 103 CFU. The numbers of CFU per organ in spleen (A, C) and liver (B, D) of individual mice are shown. Bars indicate the mean values of the experimental groups. Statistical analysis of results was performed with the Tukey multiple comparison test (p < 0.05), and asterisks indicate significantly attenuated mutants. The experimental groups were ranked according to the bacterial load. The experiments were repeated twice with similar results.
The strains EGDΔlmo2734, EGDΔlmo1085 and EGDΔlmo618 also showed a minor reduction of bacterial counts, which was more pronounced in the spleen (Figure 4C) than in the liver (Figure 4D). This trend to reduced bacterial loads, however, was not statistically significant. In the case of the remaining strains EGDΔoppF, EGDΔlmo2781 and EGDΔtagB, no reduction of bacterial counts compared to mice infected with strain EGD was observed (Figure 4A, C), although oppF is induced during mouse infection [23].
Discussion
Recently, two independent studies have disclosed the expression profile of L. monocytogenes during infection of Caco-2 cells and macrophages [11,12]. However, the up- or down-regulation of a gene under certain conditions does not always correlate with a phenotype of the respective mutant, possibly due to the high number of parameters changed, physiological side effects, catabolite repression, or transcriptional activities without impact. To overcome these limitations, we decided to complement our knowledge of listerial intracellular behaviour by the screening of a comprehensive mutant library, and report the results of a search for L. monocytogenes genes required during infection of and replication in human epithelial cells. This experimental genetic screen under defining conditions was finally combined with bioinformatic modelling.
Surface proteins
Ami, Vip, and InlA are well known virulence factors that contribute to listerial adhesion to and invasion of eukaryotic cells, and there identification validates the screening procedure. Ami is an autolysin amidase involved in adhesion to epithelial cells [24], and InlA and Vip are required for entry into several non-phagocytic eukaryotic cells [25,26]. Besides InlA and Vip, four hypothetical cell surface proteins with LPXTG motif have been identified in this screening, namely Lmo1666, and the internalin-like proteins Lmo0514, Lmo2026, and Lmo0327 [3,27-30]. A lack of these four proteins affected the capability of strain EGD to invade Caco-2 cells only slightly, but a tissue-specific phenotype of these mutants as recently shown for Lmo2026 that plays a role during listerial multiplication in the brain cannot be excluded [31]. An lmo0441 mutant has been shown to grow wildtype-like in J774 macrophages and, in contrast to our data, also in Caco-2-derived C2Bbe1 cells [32].
Virulence-related factors
Several of the genes that play a role intracellular replication as identified here are also known to be required for virulence of L. monocytogenes or other pathogens in mice. Upon application of signature-tagged transposon mutagenesis, a homologue of agrA encoding a response regulator involved in Staphylococcus aureus virulence [33], has recently been identified as virulence factor of L. monocytogenes, because its mutation resulted in a ten-fold increase in the 50% lethal dosis [34]. In contrast to this study, IDM inactivation of agrA resulted in a 3.4-fold attenuation of L. monocytogenes in Caco-2 cells, a discrepancy that might be explained by experimental conditions such as the MOI used. The inactivation of sipX (lmo1269) encoding a type I signal peptidase resulted in an eight-fold reduction of the intracellular multiplication of L. monocytogenes (data not shown) and also significantly affected the listerial virulence [35]. A deletion of fur responsible for oxidative stress response and iron storage led to a strong reduction of listerial virulence, but did not affect the growth of strain EGD-e in macrophages [36]. Our data clearly show the requirement of IspE for L. monocytogenes virulence, thus confirming a recent study that investigated mutants in other isoprenoid synthesis genes to demonstrate an in vivo role of the MEP pathway [37]. Listerial IspE exhibits a 62% similarity to IspE of Bacillus cereus and is highly conserved among L. monocytogenes strains. In contrast to our infection studies with Caco-2 cells (Table 1), isoprenoid biosynthesis is not required for listerial growth in macrophages [37]. Interestingly, IspE has been described as a novel protein target that elicits a strong antibody response from antiserum from rabbits infected with L. monocytogenes, suggesting that this protein is induced or upregulated during infection [38]. Three genes belonging to the category of membrane bioenergetics, namely lmo0091 and atpA/atpB, encode subunits of two ATP synthases for which a role in virulence has been demonstrated in Francisella novicida [39]. lmo2694, encoding a lysine decarboxylase, probably contributes to acid resistance by consuming intracellular protons [40].
Transporters
A surprisingly high number of genes involved in various transport processes contribute to the intracellular replication of L. monocytogenes, justifying their consideration in the metabolism model (Figure 3). Obviously, the eukaryotic cytosol is exploited by the bacterium for osmoprotectants and nutrients such as sugars, phosphate and amino acids. This is in line with the assumption that within cells, L. monocytogenes utilizes sugars besides phosphorylated glucose, and intracellular peptides as a source of amino acids [7]. Mutant EGDΔlmo135-137 lacking the transporter Lmo0135-0137 associated with cysteine transport is growth attenuated in Caco-2 cells. This finding is in line with the assumption that the uptake of amino acids such as alanine, aspartate and glutamate from the host cell is a requirement for intracellular replicating of L. monocytogenes [41]. A similar observation has been made for OppABCDF essential for oligopeptide uptake. An oppA mutant showed retarded growth in macrophages and affects growth at low temperature [15]. lmo0135 (ctpA), but not OppABCDF, is essential for full virulence of L. monocytogenes in mice [4], suggesting a lack of available cysteine during systemic infection. In contrast to the oligopeptide-binding protein OppA of L. monocytogenes [15], Lmo0135-0137 does not mediate the transport of bialaphos, because strain EGDΔlmo0135-0137 showed a wildtype-like susceptibility of this toxic tripeptide (data not shown). Four other loci in the genome of L. monocytogenes are predicted to be involved in (oligo)peptide uptake, namely dtpT, lmo1265, lmo1712, and lmo0152 upregulated in macrophages [11], but the activity of only two of them has been disclosed [42-44].
The requirement of lmo2430 involved in ferrichrome transport, as well as of the iron uptake regulator Fur, points to a restriction of iron availability inside the cytosol, a finding that has also been reported for Shigella flexneri during intramacrophagic growth [45]. Within epithelial cells, L. monocytogenes competes for phosphate as shown by the identification of lmo2249 encoding a low-affinity inorganic phosphate transporter. The uptake of glycine betaine by gbuA- and opuCA-encoded transporters [46] contributes to osmotolerance of L. monocytogenes and thus to intracellular proliferation.
Metabolism
Four genes involved in the synthesis of purines (purA, purQ, lmo1771) and pyrimidines (pyrE) are required for intracellular proliferation of L. monocytogenes, suggesting that these bases and nucleotides are not provided by the host cell, but must be synthesized by the bacterium. Two further genes identified here, pdxK and lmo2102, are involved in the biosynthesis of pyridoxine, a vitamin that contributes to transaminase activities during amino acid degradation. This observation also stresses that amino acid metabolism plays a key role for intracellular replication of L. monocytogenes, a finding that is supported by our network analysis of key flux modes for intracellular survival. A mutation in nadB demonstrate the requirement of nicotinate and nicotinamide metabolism during multiplication in Caco-2 cells, although a niacin-auxotrophic mutant of L. monocytogenes revealed no growth deficiencies following macrophage infection [7].
With respect to amino acid metabolism, the genes listed in Table 1, and the corresponding flux mode analysis (see Additional file 4 for results obtained from the knockout in silico experiment), show that besides the de novo biosynthesis of all aromatic and branched chain amino acids and arginine [7,12], an intact metabolism of methionine (lmo0594, lmo1495) and serine (lmo1235, serC) is required for multiplication of L. monocytogenes within Caco-2 cells.
The identification of lmo0517 and lmo2831 that encode a phosphoglycerate mutase and a phosphoglucomutase, respectively, support the finding that the pentose phosphate cycle rather than glycolysis is the predominant pathway of sugar metabolism of L. monocytogenes during proliferation in epithelial cells [12].
Conclusion
An important outcome of the systems biology approach described here is the fact that L. monocytogenes overcomes several nutrient limitations within the epithelial cytosol by the expression of genes mainly involved in transport processes and in the metabolism of sugars, cofactors and nucleic acids. Although this pathogen is assumed to encounter a nutrient-rich surrounding after escape from the phagosome, it requires a surprisingly high number of metabolic pathways and factors during intracytosolic replication. Their identity became apparent by combining a genetic screen with flux mode calculations. As a result of this metabolic modelling, we could show that Listeria pathways for intracytoplasmatic survival are clearly distinct from the central set of genes essential for survival under optimal metabolic conditions, e.g. in full medium. Certain metabolic capabilities revealed to be important for intracellular survival, while the respective mutants did not show a phenotype in full medium. The results obtained are in line with the assumption that intracellular bacteria avoid the competition with the substrate requirements of their host cell, but prefer to use excess, storage, or garbage products of the cytosol [2]. Only few mutants identified in this screen were also attenuated in the mouse infection model, indicating that L. monocytogenes uses a huge number of partially redundant pathways and nutrient acquisition strategies, all of which contribute to its highly physiological flexibility within in vivo compartments.
Methods
Bacterial strains, growth conditions, cell lines, and mice
Strains used in this study are listed in Table 2. Escherichia coli strains XL2-blue (Stratagene, La Jolla, CA) and DH5α were cultivated in Luria-Bertani (LB) medium at 37°C. L. monocytogenes EGD (serovar 1/2a) was grown at 37°C in Brain Heart Infusion (BHI) or in modified minimal Welshimer's broth (mMWB) with 0.1 g histidine per liter [47]. The temperature applied was 30°C or 43°C in the presence of temperature-sensitive plasmid pLSV101. When necessary, media were supplemented with erythromycin (Serva, Electrophoresis GmbH, Heidelberg, Germany) to a final concentration of 300 μg/ml for E. coli or 5 μg/ml for L. monocytogenes. For solid media, 1.5% agar (w/v) was added. When appropriate, medium osmolarity was increased by the addition of 3% NaCl. Human colon epithelial cells (Caco-2 cells) were received from the American Type Culture Collection (ATCC HTB-37) and were cultured at 37°C with 5% CO2 in RPMI 1640 (Biochrom KG, Berlin, Germany) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Perbio Science, Bonn, Germany). In order to monitor the growth of insertion mutants, overnight cultures were diluted 1:1000 in 250 μl BHI medium with erythromycin and shaken in a microtitre plate at 43°C for 24 h. The optical density of the cultures at 600 nm (OD600) was measured every 30 minutes in a colorimeter (Bioscreen C, Labsystem, France). For growth curves of deletion mutants, strains were grown overnight in the appropriate medium at 37°C, diluted as specified and shaken at 180 rpm in flasks until reaching stationary phase. The OD600 was measured each hour. Female BALB/c (H-2d) mice were purchased from Janvier (Le Geneste St. Isle, France), kept under conventional conditions, and used at 8-10 weeks of age.
Table 2.
Strains and plasmids used in this study
| name | characterization | reference |
|---|---|---|
| XL2-blue | E. coli: recA1, endA1, gyrA96, thi-1, hsdR17, supE44, relA1, lac, [F', proAB, laclqZΔM15Tn10(Tetr), Amy, Camr] | Stratagene |
| DH5α | E. coli: deoR, endA1, gyrA96, hsdR17(rk-mk+),recA1, relA1, supE44, λlthi-1, Δ(lacZYA-argFV169) | [59] |
| EGD | L. monocytogenes Sv 1/2a, wildtype | S. H. E. Kaufmann |
| Pkp1 | L. monocytogenes Sv 1/2a, ΔplcA/hly/mpl/actA/plcB, Kanr | [60] |
| EGDΔlmo0135-137 | in-frame deletion mutant of a putative oligonucleotide transporter gene | This study |
| EGDΔispE | in-frame deletion mutant of lmo0190 involved in mevalonate biosynthesis | This study |
| EGDΔlmo0618 | in-frame deletion mutant of a protein kinase gene | This study |
| EGDΔlmo1031-1036 | In-frame deletion mutant of an operon responsible for glycerol metabolism | This study |
| EGDΔlmo1085 | in-frame deletion mutant of lmo1085 involved in teichoic synthesis | This study |
| EGDΔtagB | in-frame deletion mutant of tagB (lmo1088) involved in teichoic synthesis | This study |
| EGDΔlmo1506 | in-frame deletion mutant of a putative transporter gene | This study |
| EGDΔoppF | in-frame deletion mutant of the putative oligopeptide ABC transporter gene lmo2192 | This study |
| EGDΔlmo2734 | in-frame deletion mutant of lmo2734 encoding a putative sugar hydrolase | This study |
| EGDΔlmo2781 | in-frame deletion mutant of lmo2781 involved in cellobiose metabolism | This study |
| pLSV101 | Temperature-sensitive shuttle vector; EmR | [12] |
| EGDΔpdxK | in-frame deletion mutant of pdxK (lmo0662) involved in biosynthesis of pyridoxine | This study |
General techniques
DNA manipulations and isolation of chromosomal DNA were performed according to standard protocols [48], and following the manufacturer's instructions. GeneRuler™ DNA Ladder Mix from MBI Fermentas (St. Leon-Rot, Germany) was used as a marker for DNA analysis. Plasmid DNA was transformed via electroporation by using a Bio-Rad Gene pulser II as recommended by the manufacturer. Polymerase chain reactions (PCRs) were carried out with Taq polymerase. Chromosomal DNA or an aliquot of a single colony resuspended in 100 μl H2O served as template for PCR. Listerial gene annotation was done according to the Listeria homepage of the Institut Pasteur http://genolist.pasteur.fr/ListiList/, and the oligonucleotides used for PCRs are listed in Additional file 5.
Construction of a mutant library of L. monocytogenes EGD
A random mutant library of L. monocytogenes strain EGD Pkp1 was established as described recently [12]. Strain Pkp1 characterised by deletions of plcA, hly, mpl, actA, and plcB was chosen to avoid repeated identification of these well-known virulence genes. Briefly, chromosomal DNA was sonified, and the fractionated DNA was digested with MboI. Following separation by agarose gel electrophoresis, gel slices containing unirradiated DNA fragments from 200 to 400 bp were isolated. Purified chromosomal DNA was then ligated to MboI-restricted pLSV101. Sixteen independent ligation samples were transformed into E. coli XL2-blue, and a total of 3658 E. coli transformants was selected at 37°C on LB agar containing erythromycin. The average length of the cloned chromosomal fragments was 244 (+/-102) bp as determined by sequencing of 76 E. coli clones. Colonies were pooled in sets of 60-380 clones, and 300 ng DNA isolated from each pool was transformed into L. monocytogenes EGD. 3648 L. monocytogenes EGD fragment library clones were selected at 30°C in the presence of erythromycin, and single colonies were suspended in 200 μl BHI in a 96-well microtitre plate. 20 μl of each suspension were dropped with a multichannel pipette on BHI agar plates containing erythromycin and incubated at 43°C for two days [49]. Illegitimate insertion of pLSV101 without fragment was not observed. 1491 insertion mutants of L. monocytogenes EGD were isolated in 96-well microtitre plates in BHI and regrown over night at 43°C in the presence of erythromycin. Excision of the plasmid from the chromosomal site of insertion was obtained by repeated passage of a mutant clone on agar plates without erythromycin at 30°C, and the sites of plasmid insertion were determined by sequencing of plasmid-borne fragments derived from PCR with the primer pair LSV3 and LSV-4380rev [12]. Given a redundancy of 15% of the fragment library according to sequencing results, the 1491 clones tested here represent approximately 32% of the genome of strain L. monocytogenes EGD Pkp1 [12,50] and probably a higher percentage of functions encoded by operons. The stability of chromosomal pLSV101 integration during cell culture assays had already been demonstrated [12].
Construction of deletion mutants
In-frame deletions of ten genes or gene loci were performed in the parental strains Sv1/2a EGD, namely of lmo1031-1036, lmo0135-0137, ispE (lmo190), lmo0618, tagB (lmo1088), lmo1085, lmo1506, lmo2192, lmo2734, and lmo2781. The standard procedure has recently been described [12,16] and is exemplified here for the deletion of lmo0190. Two fragments of approximately 500 bp were amplified from chromosomal DNA of strain EGD using the oligonucleotide pairs Lmo0189A/Lmo0189B and Lmo0191C/Lmo0191D, and then ligated via the introduced BglII sites. Following nested PCR using the oligonucleotides Lmo0189NestedAB and Lmo0191NestedCD and the ligation mixture as a template, the resulting fragment was cloned into pLSV101 via BamHI and EcoRI giving rise to pKS0190del. pKS0190del was then transformed into L. monocytogenes EGD by electroporation, and erythromycin-resistant bacteria growing at 43°C harbouring the chromosomally integrated plasmid were selected. Cointegrates were resolved by reiterated growth at 30°C, and erythromycin-sensitive clones were screened by PCR to identify the lmo0190 (ispE) deletion mutant. The gene deletions in all ten mutants listed in Table 2 were confirmed by sequencing. All primers are indicated in the Additional file 5.
Caco-2 cell infection assays
Caco-2 cells (2.5 × 105 per well) were seeded in a 24-well culture plate and cultivated 22 h until infection. Cells were washed twice with Mg2+-and Ca2+-containing phosphate-buffered saline (PBS/Mg2+Ca2+) and covered for 1 h with 500 μl RPMI 1640 containing 2.0 μl of a bacterial culture grown over night. The average multiplicity of infection (MOI) was calculated to range from 6 to 14. To test deletion mutants, strains were grown in 20 ml BHI to late log-phase (OD600~1.0); aliquots were supplemented with glycerol at a final concentration of 15% and frozen at -80°C. Prior to infection, glycerol stocks were thawed, and the bacteria were sedimented and washed twice with PBS. After resuspension in 1 ml RPMI 1640, the number of viable bacteria was determined as CFU. The average MOI used here was 8 to 11.
After an infection period of 1 h, the Caco-2 cells were washed twice with PBS/Mg2+Ca2+. Extracellular bacteria were removed by adding 0.5 ml RPMI 1640 with 50 μg/ml gentamicin for 1 h, and the medium was then replaced by RPMI 1640 with 10 μg/ml gentamicin. At appropriate time points of incubation (2 h and 8 h), the infected cells were washed with PBS/Mg2+Ca2+ and then lysed in 1 ml cold Triton X-100 (0.1%). Intracellular replication behaviour of the mutants and the wildtype was quantified by plating dilutions of the lysed cells on BHI agar plates that were incubated at 37°C and 43°C, respectively, for one day. If appropriate, the plates contained 5 μg/ml erythromycin. To examine adhesion properties of bacterial strains, the infection time was reduced to 35 min, and before lysis, cells were washed four times with PBS/Mg2+Ca2+. The capability of bacterial cells to invade Caco-2 cells was investigated as described above, but lysis of the epithelial cells was performed after 1 h, and a higher gentamicin concentration of 50 μg/ml was used. In all experiments, intact eukaryotic cell monolayers were observed prior to cell lysis.
Mice infection assays
Female BALB/c (H-2d) mice were purchased (Janvier, Le Geneste St. Isle, France), kept under conventional conditions, and used at 8-10 weeks of age. Animal experiments were approved according to German federal law (Baden-Württemberg, permission number G-3/06). Mice were infected intravenously via the tail vein with a sublethal dose of L. monocytogenes serovar 1/2a EGD or L. monocytogenes-derived mutants in 0.2 ml endotoxin-free phosphate-buffered saline as indicated. Bacteria used for infection were in the logarithmic growth phase. The bacterial concentration of inoculated bacteria was calculated from OD600 and confirmed by plating on blood agar. Liver and spleen were removed three and six days after infection, respectively. The number of CFU per organ homogenate was determined by pour-plating dilutions of organ homogenates in BHI agar. The detection limit of the assay was 100 colony forming units (CFU) per organ. The statistical significance of the results of the mouse infection experiments was analyzed with the Tukey multiple comparison test [51] at the 0.05 significance level after logarithmic transformation of CFU and PFU values. This test analyzes the significance of the difference between all possible pairs of means with appropriate adjustment for the multiple testing. Calculations were performed using the WINKS statistical analysis software (TEXASOFT, Cedar Hill, USA).
Modelling
The genome-scale metabolic network was reconstructed according to the latest annotation of L. monocytogenes EGD-e (GenBank accession number: NC_003210) based on the EGD-e genome sequence [13]. Reconstruction was done by systematic genome comparisons applying InGeno [52] and extensive sequence and domain analysis [53]. Metabolite terms and the topological structure were taken from the KEGG metabolic database. The topological structures were visualized and revised using the YANA and YANA square software [54,55]. A condensed network consisting of 167 metabolites, 155 enzymes and transporters was used to model the basic listerial metabolism based in a genome scale. Various carbon sources such as glucose, fucose, glycerol, acetate and citrate were considered. All essential amino acids are assumed to be synthesized de novo in the presence of sufficient amounts of ammonia. The null-space calculation from the convex basis resulted in 163 extreme pathways (EPs) and we calculated 20826 elementary modes (EMs) for the network.
Abbreviations
MOI: multiplicity of infection; IDM: insertion duplication mutagenesis
Authors' contributions
KS performed most experimental work with Listeria including infection assays and mutant construction, GG was responsible for the mice infection studies and CL for the modelling, WG and TD contributed to the conception and revised the manuscript, TMF designed and coordinated the study, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Literature data regarding genes affecting intracellular replication of L. monocytogenes.
Selected genes used for modelling of the listerial metabolism during intracellular replication.
Input data of the model for calculation with flux balance analysis (format: Suitable for Metatool or YANAsquare; Schwarz et al., 2007) including major transporters, amino acids and intermediary metabolites as shown in Figure 4. The model allows also further detailed analyses, e.g. of subnetworks as well as metabolic fluxes for identifying essential genes under different physiological growth conditions, such as medium or intracellular Caco-2 (PERL scripts were used for calculating different subsets).
Results obtained from the knockout in silico experiment are summarized in this document (Tables S1-S3, Figure S1). Table S1: Metabolic flux modes calculated using extreme pathway analysis. Table S2: Key enzymes occurring in all the flux modes. Table S3: Metabolic flux modes critical for cytoplasmic survival interpreted as pathway equations. Figure S1: Elementary mode numbers affected by cytoplasmically attenuated mutants.
Oligonucleotides used in this study.
Total cell number of mutants after invasion and after 7 hours of intracellular replication; the generation time is indicated.
Contributor Information
Kristina Schauer, Email: kristina.schauer@wzw.tum.de.
Gernot Geginat, Email: gernot@geginat.umm.de.
Chunguang Liang, Email: Liang@biozentrum.uni-wuerzburg.de.
Werner Goebel, Email: goebel@mvp.uni-muenchen.de.
Thomas Dandekar, Email: Dandekar@biozentrum.uni-wuerzburg.de.
Thilo M Fuchs, Email: thilo.fuchs@wzw.tum.de.
Acknowledgements
The authors thank Samir Velagic for technical assistance. This work was supported by the Competence Center PathoGenoMik funded by the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF), Germany, and by the Priority Program SPP 1316 of the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG; Fu 375/5-1; Da 208/10-1;11-1).
References
- Junttila JR, Niemela SI, Hirn J. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J Appl Bacteriol. 1988;65:321–327. doi: 10.1111/j.1365-2672.1988.tb01898.x. [DOI] [PubMed] [Google Scholar]
- Joseph B, Goebel W. Life of Listeria monocytogenes in the host cells' cytosol. Microbes Infect. 2007;9:1188–1195. doi: 10.1016/j.micinf.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- Port GC, Freitag NE. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect Immun. 2007;75:5886–5897. doi: 10.1128/IAI.00845-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain T, Chatterjee SS, Ghai R, Kuenne CT, Billion A, Steinweg C, Domann E, Karst U, Jansch L, Wehland J, Eisenreich W, Bacher A, Joseph B, Schar J, Kreft J, Klumpp J, Loessner MJ, Dorscht J, Neuhaus K, Fuchs TM, Scherer S, Doumith M, Jacquet C, Martin P, Cossart P, Rusniock C, Glaser P, Buchrieser C, Goebel W, Chakraborty T. Pathogenomics of Listeria spp. Int J Med Microbiol. 2007;297:541–557. doi: 10.1016/j.ijmm.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Carbon metabolism of intracellular bacteria. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- Marquis H, Bouwer HG, Hinrichs DJ, Portnoy DA. Intracytoplasmic growth and virulence of Listeria monocytogenes auxotrophic mutants. Infect Immun. 1993;61:3756–3760. doi: 10.1128/iai.61.9.3756-3760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld AD, Goossens PL, Cossart P. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol Microbiol. 1994;13:585–597. doi: 10.1111/j.1365-2958.1994.tb00453.x. [DOI] [PubMed] [Google Scholar]
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, Vazquez-Boland JA. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci USA. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan M, Moors MA, Portnoy DA. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. Intracellular Gene Expression Profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Garcia-del Portillo F, Garrido P, Gautier L, Goebel W, Gomez-Lopez N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Perez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vazquez-Boland JA, Voss H, Wehland J, Cossart P. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- Eisenreich W, Slaghuis J, Laupitz R, Bussemer J, Stritzker J, Schwarz C, Schwarz R, Dandekar T, Goebel W, Bacher A. 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc Natl Acad Sci USA. 2006;103:2040–2045. doi: 10.1073/pnas.0507580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borezee E, Pellegrini E, Berche P. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun. 2000;68:7069–7077. doi: 10.1128/IAI.68.12.7069-7077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Stolz J, Scherer S, Fuchs TM. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J Bacteriol. 2009;191:2218–2227. doi: 10.1128/JB.01636-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol Med Microbiol. 2003;35:207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- Fuchs TM, Klumpp J, Przybilla K. Insertion-duplication mutagenesis of Salmonella enterica and related species using a novel thermosensitive vector. Plasmid. 2006;55:39–49. doi: 10.1016/j.plasmid.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Joseph B, Mertins S, Stoll R, Schar J, Umesha KR, Luo Q, Muller-Altrock S, Goebel W. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin JA, Stelling J, Price ND, Klamt S, Schuster S, Palsson BO. Comparison of network-based pathway analysis methods. Trends Biotechnol. 2004;22:400–405. doi: 10.1016/j.tibtech.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Begley M, Gahan CG, Kollas AK, Hintz M, Hill C, Jomaa H, Eberl M. The interplay between classical and alternative isoprenoid biosynthesis controls γδ T cell bioactivity of Listeria monocytogenes. FEBS Lett. 2004;561:99–104. doi: 10.1016/S0014-5793(04)00131-0. [DOI] [PubMed] [Google Scholar]
- Xayarath B, Marquis H, Port GC, Freitag NE. Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol Microbiol. 2009;74:956–973. doi: 10.1111/j.1365-2958.2009.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camejo A, Buchrieser C, Couve E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009;5:e1000449. doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milohanic E, Pron B, Berche P, Gaillard JL. Identification of new loci involved in adhesion of Listeria monocytogenes to eukaryotic cells. European Listeria Genome Consortium. Microbiology. 2000;146:731–739. doi: 10.1099/00221287-146-3-731. [DOI] [PubMed] [Google Scholar]
- Cabanes D, Sousa S, Cebria A, Lecuit M, Garcia-del Portillo F, Cossart P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. Embo J. 2005;24:2827–2838. doi: 10.1038/sj.emboj.7600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-N. [DOI] [PubMed] [Google Scholar]
- Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002;10:238–245. doi: 10.1016/S0966-842X(02)02342-9. [DOI] [PubMed] [Google Scholar]
- McGann P, Ivanek R, Wiedmann M, Boor KJ. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae. Appl Environ Microbiol. 2007;73:2806–2814. doi: 10.1128/AEM.02923-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowska M, Markiewicz Z. Characterization of Listeria monocytogenes protein Lmo0327 with murein hydrolase activity. Arch Microbiol. 2006;186:69–86. doi: 10.1007/s00203-006-0122-8. [DOI] [PubMed] [Google Scholar]
- Bierne H, Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol Mol Biol Rev. 2007;71:377–397. doi: 10.1128/MMBR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Luo X, Jiang L, Jin P, Wei W, Liu D, Fang W. Molecular characteristics and virulence potential of Listeria monocytogenes isolates from Chinese food systems. Food Microbiol. 2009;26:103–111. doi: 10.1016/j.fm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Guinane CM, Cotter PD, Ross RP, Hill C. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob Agents Chemother. 2006;50:2824–2828. doi: 10.1128/AAC.00167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autret N, Dubail I, Trieu-Cuot P, Berche P, Charbit A. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect Immun. 2001;69:2054–2065. doi: 10.1128/IAI.69.4.2054-2065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnemain C, Raynaud C, Reglier-Poupet H, Dubail I, Frehel C, Lety MA, Berche P, Charbit A. Differential roles of multiple signal peptidases in the virulence of Listeria monocytogenes. Mol Microbiol. 2004;51:1251–1266. doi: 10.1111/j.1365-2958.2004.03916.x. [DOI] [PubMed] [Google Scholar]
- Rea RB, Gahan CG, Hill C. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect Immun. 2004;72:717–727. doi: 10.1128/IAI.72.2.717-727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Bron PA, Heuston S, Casey PG, Englert N, Wiesner J, Jomaa H, Gahan CG, Hill C. Analysis of the isoprenoid biosynthesis pathways in Listeria monocytogenes reveals a role for the alternative 2-C-methyl-D-erythritol 4-phosphate pathway in murine infection. Infect Immun. 2008;76:5392–5401. doi: 10.1128/IAI.01376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WL, Dan H, Lin M. Novel protein targets of the humoral immune response to Listeria monocytogenes infection in rabbits. J Med Microbiol. 2007;56:888–895. doi: 10.1099/jmm.0.46977-0. [DOI] [PubMed] [Google Scholar]
- Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, Brittnacher MJ, Manoil C, Skerett SJ, Salama NR. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun. 2009;77:232–244. doi: 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW, Park YK, Bang IS, Karem K, Betts H, Hall HK, Shaw E. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology. 1994;140(Pt 2):341–352. doi: 10.1099/13500872-140-2-341. [DOI] [PubMed] [Google Scholar]
- Eylert E, Schar J, Mertins S, Stoll R, Bacher A, Goebel W, Eisenreich W. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol Microbiol. 2008;69:1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x. [DOI] [PubMed] [Google Scholar]
- Verheul A, Hagting A, Amezaga MR, Booth IR, Rombouts FM, Abee T. A di- and tripeptide transport system can supply Listeria monocytogenes Scott A with amino acids essential for growth. Appl Environ Microbiol. 1995;61:226–233. doi: 10.1128/aem.61.1.226-233.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheul A, Rombouts FM, Abee T. Utilization of oligopeptides by Listeria monocytogenes Scott A. Appl Environ Microbiol. 1998;64:1059–1065. doi: 10.1128/aem.64.3.1059-1065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters JA, Hain T, Darji A, Hufner E, Wemekamp-Kamphuis H, Chakraborty T, Abee T. Identification and characterization of Di- and tripeptide transporter DtpT of Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2005;71:5771–5778. doi: 10.1128/AEM.71.10.5771-5778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Liu H, Jin Q, Hinton JC, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun. 2005;73:88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemekamp-Kamphuis HH, Wouters JA, Sleator RD, Gahan CG, Hill C, Abee T. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl Environ Microbiol. 2002;68:4710–4716. doi: 10.1128/AEM.68.10.4710-4716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne RJ, Lin WJ, Johnson EA. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 1991;57:3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y; 2001. [Google Scholar]
- Knuth K, Niesalla H, Hueck CJ, Fuchs TM. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- Lee MS, Dougherty BA, Madeo AC, Morrison DA. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl Environ Microbiol. 1999;65:1883–1890. doi: 10.1128/aem.65.5.1883-1890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. Exploratory Data Analysis. Reading, Ma: Addison-Wesley; 1977. [Google Scholar]
- Liang C, Dandekar T. inGeno-an integrated genome and ortholog viewer for improved genome to genome comparisons. BMC Bioinformatics. 2006;7:461. doi: 10.1186/1471-2105-7-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudermann P, Vogl I, Zientz E, Silva FJ, Moya A, Gross R, Dandekar T. Analysis of and function predictions for previously conserved hypothetical or putative proteins in Blochmannia floridanus. BMC Microbiol. 2006;6:1. doi: 10.1186/1471-2180-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Musch P, von Kamp A, Engels B, Schirmer H, Schuster S, Dandekar T. YANA - a software tool for analyzing flux modes, gene-expression and enzyme activities. BMC Bioinformatics. 2005;6:135. doi: 10.1186/1471-2105-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Liang C, Kaleta C, Kuhnel M, Hoffmann E, Kuznetsov S, Hecker M, Griffiths G, Schuster S, Dandekar T. Integrated network reconstruction, visualization and analysis using YANAsquare. BMC Bioinformatics. 2007;8:313. doi: 10.1186/1471-2105-8-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll R, Mertins S, Joseph B, Muller-Altrock S, Goebel W. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology. 2008;154:3856–3876. doi: 10.1099/mic.0.2008/018283-0. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Engelbrecht F, Chun SK, Ochs C, Hess J, Lottspeich F, Goebel W, Sokolovic Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Literature data regarding genes affecting intracellular replication of L. monocytogenes.
Selected genes used for modelling of the listerial metabolism during intracellular replication.
Input data of the model for calculation with flux balance analysis (format: Suitable for Metatool or YANAsquare; Schwarz et al., 2007) including major transporters, amino acids and intermediary metabolites as shown in Figure 4. The model allows also further detailed analyses, e.g. of subnetworks as well as metabolic fluxes for identifying essential genes under different physiological growth conditions, such as medium or intracellular Caco-2 (PERL scripts were used for calculating different subsets).
Results obtained from the knockout in silico experiment are summarized in this document (Tables S1-S3, Figure S1). Table S1: Metabolic flux modes calculated using extreme pathway analysis. Table S2: Key enzymes occurring in all the flux modes. Table S3: Metabolic flux modes critical for cytoplasmic survival interpreted as pathway equations. Figure S1: Elementary mode numbers affected by cytoplasmically attenuated mutants.
Oligonucleotides used in this study.
Total cell number of mutants after invasion and after 7 hours of intracellular replication; the generation time is indicated.