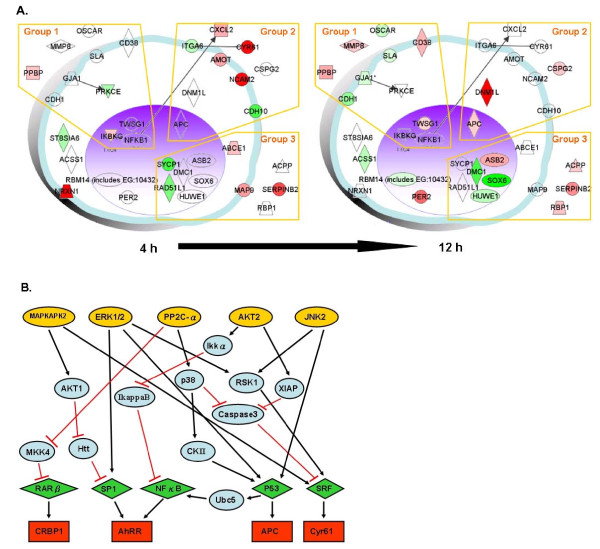
Figure 3.
Pathway analysis of representative genes that responded to [BF/S+L/Ep] treatment. A prototypical cell was constructed from 37 representative genes that responded to treatment with [BF/S+L/Ep] in vitro from 4 h to 12 h. A, Genes whose expression was up-regulated (more than doubled) are colored red, and those whose expression was down-regulated (to less than half) are shown in green. Selected regions of the network highlight several groups of genes. Group 1, Immune response related genes. Group 2, Adhesion molecules and cytoskeleton; cell movement related genes. Group 3, Cell cycle, cell proliferation and apoptosis related genes. Gene networks were analyzed with the Ingenuity Pathways program. B, The TRANSPATH Professional 7.1 database was searched to assess possible signaling pathways, networks or potential interactions among the responsive genes/target molecules in DCs treated with [BF/S+L/Ep]. The 37 genes that were up- or down-regulated at least 2-fold compared to controls were analyzed. Specifically, connections (hits) within 7 genes were employed as the parameter for the current search. Five postulated key molecules/pathways; JNK2 (c-Jun NH2-terminal kinase 2), PP2C-α (protein phosphatase 2C-alpha), AKT (protein kinase B), ERK (extracellular signal-related kinase) and MAPKAPK (MAPK-activated protein kinase 2) that may be activated by [BF/S+L/Ep] treatment are predicted with arrows. The green rhombuses indicate the upstream transcription factors of responsive genes.