Summary
Magnetotactic bacteria contain nanometer-sized, membrane-bound organelles, called magnetosomes, which are tasked with the biomineralization of small crystals of the iron oxide magnetite allowing the organism to use geomagnetic field lines for navigation. A key player in this process is the HtrA/DegP family protease MamE. In its absence, Magnetospirillum magneticum str AMB-1 is able to form magnetosome membranes but not magnetite crystals, a defect previously linked to the mislocalization of magnetosome proteins. In this work we use a directed genetic approach to find that MamE, and another predicted magnetosome-associated protease, MamO, likely function as proteases in vivo. However, as opposed to the complete loss of mamE where no biomineralization is observed, the protease-deficient variant of this protein still supports the initiation and formation of small, 20 nm-sized crystals of magnetite, too small to hold a permanent magnetic dipole moment. This analysis also reveals that MamE is a bifunctional protein with a protease-independent role in magnetosome protein localization and a protease-dependent role in maturation of small magnetite crystals. Together these results imply the existence of a previously unrecognized “checkpoint” in biomineralization where MamE moderates the completion of magnetite formation and thus committal to magneto-aerotaxis as the organism’s dominant mode of navigating the environment.
Keywords: bacterial organelle, HtrA/DegP protease, magnetosome, magnetite, biomineralization, heme-binding
Introduction
Organelles have long been hailed as a defining feature of eukaryotic cells, a definition that is changing as we gain more insight into the membrane- and protein-bound compartments found in bacteria and archaea (Shively, 2006, Fuerst, 2005, Murat et al., 2010a). The existence of organelles in bacteria presents interesting evolutionary questions about the conservation of mechanisms of organelle formation, maintenance and organization. Bacterial organelles have also received attention because they often compartmentalize otherwise inefficient or toxic processes, a feature which may be exploited to carry out useful, but potentially cytotoxic, reactions.
One such organelle is the magnetosome of magnetotactic bacteria, which facilitates biomineralization processes that in vitro require harsh cytotoxic conditions. The magnetosome is a membrane-bound compartment that directs biomineralization of nanometer-sized, fixed single domain crystals of iron oxide (magnetite, Fe3O4) or iron sulfide (greigite, Fe3S4). In a species-dependent manner, cells contain anywhere from ten to hundreds of magnetosomes, which are organized into one or several chains, allowing the cells to align with the earth’s magnetic field lines. This passive alignment is thought to facilitate the bacterium’s search for favored low oxygen environments, a process referred to as magnetoaerotaxis (Frankel et al., 1997, Smith et al., 2006, Komeili, 2007). The magnetosome membrane is enriched for a specific set of proteins termed magnetosome proteins (Grunberg et al., 2004, Tanaka et al., 2006). Many of these proteins, as well as other factors implicated in magnetosome formation, are encoded by a genomic island, the magnetosome island (MAI), which is a region essential for magnetosome formation conserved in all magnetotactic bacteria studied to date (Fukuda et al., 2006, Richter et al., 2007, Grunberg et al., 2001). The proteins found at the magnetosome are thought to facilitate magnetosome membrane formation, crystal formation, and chain formation and it is hypothesized that they impart the species-specific size and shape of the magnetic mineral.
In Magnetospirillum magneticum AMB-1 (AMB-1), magnetosomes are invaginations of the inner membrane formed in a step-wise fashion, where membrane invagination is followed by sorting of magnetosome proteins and magnetite biomineralization (Komeili, 2007, Murat et al., 2010b). Of particular interest is the question of how magnetotactic bacteria generate such uniformly sized and shaped crystals of magnetite, since traditional recapitulation of magnetite synthesis in vitro yields a heterogeneous mixture of crystals. How bacteria can exert such tight control over the process of magnetite formation is of interest for many proposed applications in medicine and biotechnology (Schuler & Frankel, 1999) and has driven much of the research on magnetotactic bacteria. In addition, understanding the steps and proteins involved in magnetite biomineralization may also inform how other processes of biomineralization, such as those of teeth or bone, are accomplished.
Although little is known about the molecular mechanisms underlying magnetite biomineralization, recent biochemical and genetic studies have implicated several factors in this process. For instance, addition of the magnetosome protein Mms6 to in vitro magnetite synthesis reactions confers more shape and size homogeneity, thus illustrating both the ability of magnetosome proteins to impart shape to magnetite crystals and the potential that specific proteins present for future applications (Arakaki et al., 2003, Arakaki et al., 2010). In Magnetospirillum gryphiswaldense MSR-1 (MSR-1), the absence of the mamCDFG operon results in cells that form crystals approximately 75% of the size of wildtype crystals, and it was suggested that the activity of these four genes in concert may exercise regulatory or accessory functions in crystal formation (Scheffel et al., 2008). Several factors important for magnetite biomineralization were also recently identified in a genetic dissection of the MAI in AMB-1. Confirming the importance of the mamCDFG and mms6 gene clusters, the loss of a large region of the MAI, R3, which contains both of these gene clusters, resulted in a severe defect in crystal size. In addition, other genes, including mamP, mamR, mamS and mamT, were implicated in control of crystal size, number and/or shape. Lastly, four genes, mamM, mamN, mamE and mamO, were shown to be essential for the early steps of biomineralization, as the independent deletion of each gene led to a complete absence of minerals within magnetosomes (Murat et al., 2010b).
Two of these genes, mamE and mamO encode putative HtrA/DegP family proteases. The members of this family of serine proteases are found in bacteria, archaea and eukaryotes and are known for their involvement in essential housekeeping functions such as the degradative removal of unfolded proteins during the periplasmic stress response, the initiation of the σE stress response and the removal of peroxisomal targeting signals (Kim & Kim, 2005, Clausen et al., 2002, Schuhmann et al., 2008). In AMB-1, several HtrA/DegP family proteases are encoded outside of the MAI. MamE and MamO, however, are encoded by the MAI suggesting a specific connection to magnetosome formation rather than a general role in cellular homeostasis. Accordingly, in the absence of mamE or mamO, cells are viable and form magnetosome membranes, but fail to produce minerals within these membranes (Murat et al., 2010b). This phenotype could be due to a specific defect in mineral formation or result from a more general defect in the localization of biomineralization proteins to the magnetosome. The latter was suggested for MamE since two cytoplasmic magnetosome-associated proteins are mislocalized in the mamE deletion strain (Murat et al., 2010b). Also suggestive of less canonical functions for MamE and MamO is the presence of additional functional domains (Clausen et al., 2002). MamE contains two putative c-type cytochrome CXXCH heme-binding motifs, and MamO contains a domain of unknown function (DUF81) predicted to code for seven transmembrane domains. Whether the putative protease activities of MamE and MamO are required for magnetosome formation and whether the unusual additional domains are of importance to their function was unknown. We thus undertook a mechanistic dissection of MamE and MamO to further define their roles in magnetosome biogenesis and to begin to develop an understanding of magnetosome formation at the molecular level.
Using a site directed mutagenesis, we show that MamE and MamO are likely to act as proteases in vivo and that the additional domains, uncommon for HtrA/DegP family proteases, are important for their functions. By dissecting MamE’s functional domains, we find that MamE is a bifunctional protein with a previously unidentified role in biomineralization that can be decoupled from its role in the proper localization of magnetosome proteins. Based on these results, we propose that MamE’s protease activity is required for a previously unidentified crystal size transition from 20nm crystals too small to hold a fixed magnetic dipole moment, to large crystals that can contribute to the cell’s magnetic response.
Results
MamE and MamO act as proteases in vivo
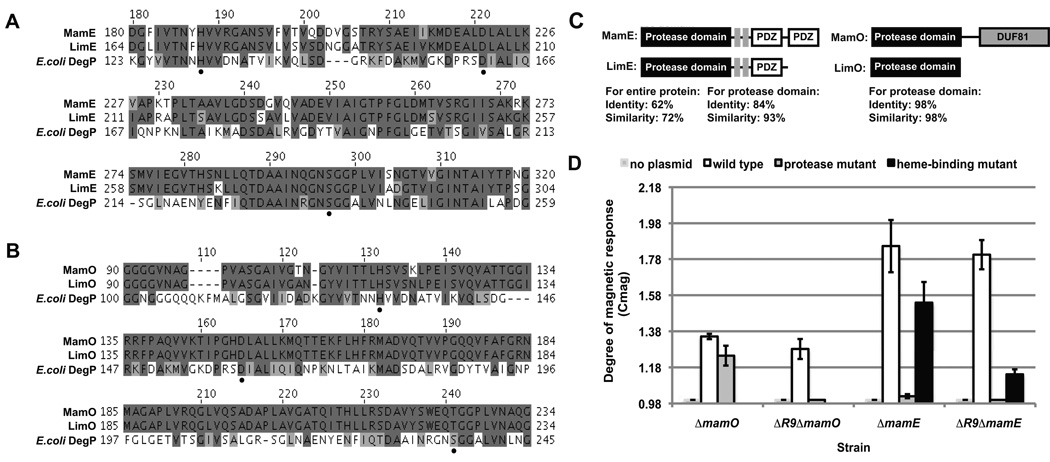
MamE and MamO are putative HtrA/DegP family proteases encoded by genes found within the MAI. In their absence, cells are non-magnetic yet still possess the ability to form magnetosome membranes (Murat et al., 2010b, Yang et al., 2010). Members of the HtrA/DegP family of proteins are trypsin-like serine proteases that share a high degree of sequence homology within their protease domains, including a highly conserved active site triad (Kim & Kim, 2005). Primary sequence alignment of MamE and MamO with E. coli DegP allowed identification of their putative active site residues. MamE shares the conserved histidine-aspartate-serine active site (Fig. 1A), whereas MamO has a threonine in place of the serine as the predicted active site nucleophile (Fig. 1B). To determine whether MamE and MamO’s putative protease functions are required for magnetite crystal formation, we generated the mutant constructs, mamEP and mamOP, by site-directed mutagenesis of the predicted active site triad residues to alanines. As a preliminary assay for the activity of these variants in vivo, we relied on the Cmag measurement, a differential spectrophotometric assay that quantifies the ability of the bacteria to orient in an external magnetic field (Schuler et al., 1995). In this assay, mamOP fully complemented the mamO deletion, and mamEP partially complemented the mamE deletion (Fig. 1D), suggesting that the putative protease functions are not essential for crystal formation.
Fig. 1. AMB-1 magnetic response (Cmag) depends on MamE and MamO protease activity.
A. Alignments of MamE and LimE protease domains with E. coli DegP.
B. Alignments of MamO and LimO protease domains with E. coli DegP. Active site triad residues of MamE and MamO, indicated by black dots, were identified based on homology with DegP. Identical residues are highlighted in dark grey and conserved substitutions in light grey.
C. Domain architecture of MamE, LimE, MamO and LimO. MamE heme-binding motifs are indicated by grey boxes. For MamE and LimE, identity for both the entire protein and for the protease domains only are shown. For MamO and LimO identity of the protease domain is shown.
D. Complementation of magnetic response of the single and double mamO and mamE deletion strains with wildtype and protease-inactive (mamEP and mamOP) constructs. For MamE, complementation by the heme-binding-deficient construct (mamEC2) is also shown. Error bars represent one standard deviation from ≥10 independent cultures.
The MAI, however, encodes paralogues of both MamE and MamO, Amb1002 and Amb1004, respectively (Figs. 1A and B) which we have renamed Like-MamE (LimE) and Like-MamO (LimO). Since HtrA/DegP proteases are known to function as oligomeric assemblies (Clausen et al., 2002, Kim & Kim, 2005) we hypothesized that these paralogues could form hetero-oligomeric complexes with MamE and MamO and thus provide active protease domains in trans. To test this hypothesis, we deleted mamE and mamO in the R9 deletion background. R9 is a region of the MAI containing both limE and limO that can be deleted in the wildtype background without a detectable phenotype (Murat et al., 2010b). The resulting double deletion strains, ΔR9ΔmamE and ΔR9ΔmamO, could be complemented to the same degree as the single deletion strains by wildtype mamE and mamO, respectively However, the putative protease-inactive MamE mutant could not restore a measurable magnetic response when expressed in the ΔR9ΔmamE strain (Fig. 1D). Constructs of MamO in which all putative active site triad residues of the protease domain are mutated to alanines also did not restore a magnetic phenotype to the ΔR9ΔmamO strain. As noted above, however, MamO has a threonine in the position of the conserved active site serine residue. This threonine can be changed to an alanine without reduction of MamO’s ability to complement ΔR9ΔmamO (Fig. S1). A single point mutation of the putative active site histidine, however, decreased MamO activity, a phenotype that would be expected if this residue were part of the serine protease active site triad (Fig. S1).
This mutational analysis suggests that the two proteins may function as proteases in vivo and that they may act within hetero-assemblies with LimE and LimO, respectively. To build on these initial results we chose to focus on MamE, with the aim of defining the specific function of its putative protease activity in magnetosome protein targeting and magnetite formation. Additionally, to avoid background effects from the unusual cross-complementation observed above, all subsequent experiments were performed in the ΔR9ΔmamE background.
MamE protease mutant produces small magnetite crystals
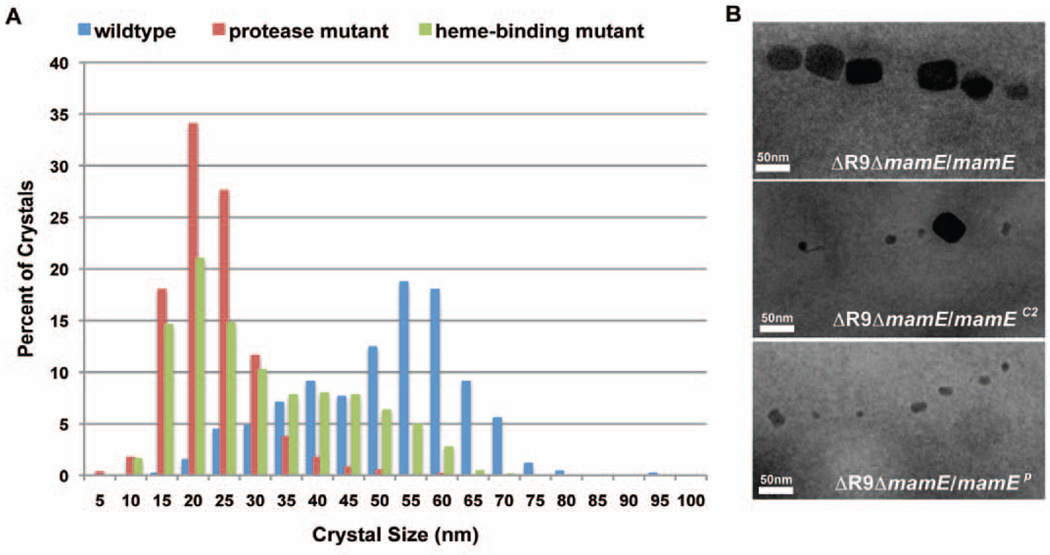
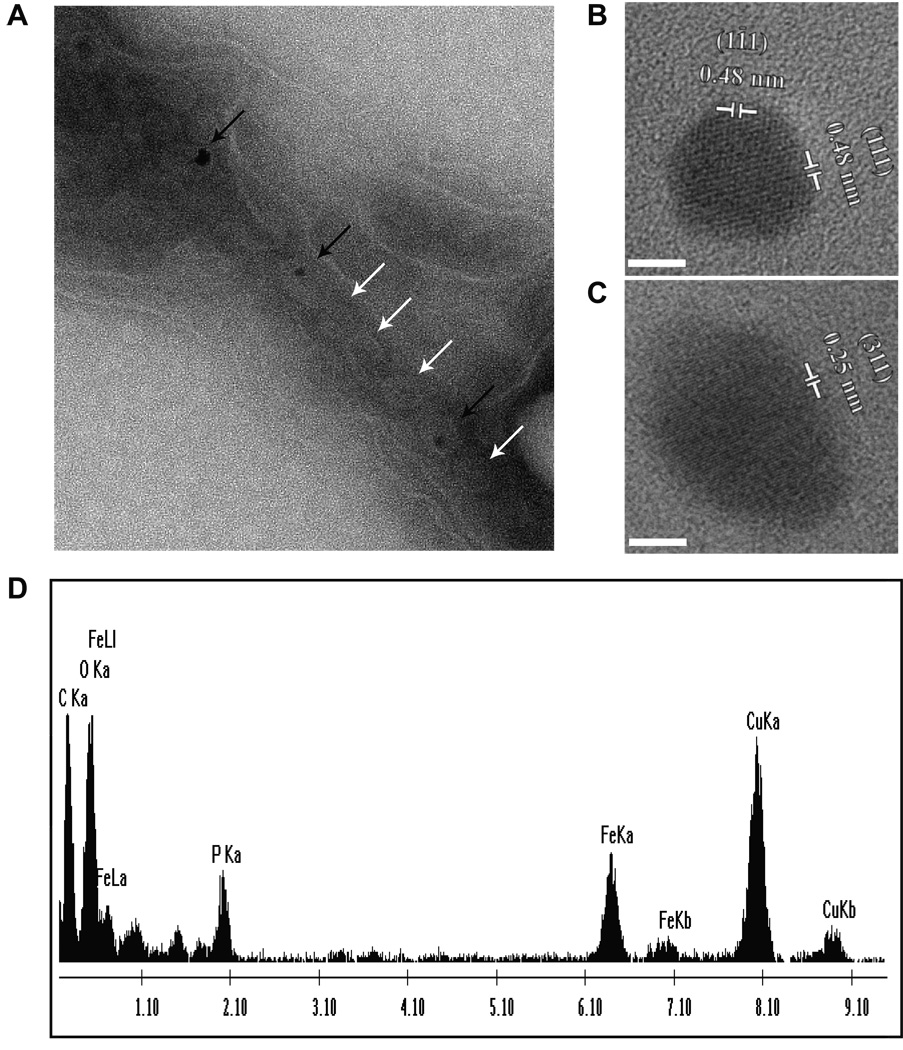
To further characterize the phenotype of the ΔR9ΔmamE/mamEP strain, cells were imaged by transmission electron microscopy (TEM). As described above, this mutant could not restore a magnetic response in the ΔR9ΔmamE strain. However, TEM analysis revealed that it still allowed for synthesis of chains of small electron dense structures (Fig 2). Under the same growth conditions, the ΔR9ΔmamE strain complemented with wildtype mamE showed a bimodal crystal size distribution with peaks centered in the 36–40 nm and 51–55 nm size ranges (Fig. 2). In contrast, when mamEP was expressed in the ΔR9ΔmamE strain, particle size distribution was centered at 16–20 nm, and only 3% were larger than 35 nm. Magnetite crystals smaller than 30 nm are too small to hold a permanent magnetic dipole moment (Butler & Banerjee, 1975), perhaps explaining the inability of the mamEP mutant to align in a magnetic field. High resolution TEM (HRTEM) confirmed that these 16–20 nm particles, and even those that were irregular in shape, were indeed crystalline (Fig. 3B and C, Fig S2). HRTEM allows for visualization of the lattice fringes of crystalline material and the spacing between these fringes can be characteristic of the crystallographic planes of various minerals. Lattice fringe images of most of the crystalline particles, formed when the mamEP mutant was expressed in the ΔR9ΔmamE strain, showed a characteristic d-spacing of 0.48 nm corresponding to {111} plane of magnetite (Figs. 3B and S2). The XRD data of magnetite in the mineral database <http://rruff.info> was used as reference for these measurements. Other crystals showed measurements that were also consistent with other planes of magnetite. In addition, spectra from energy dispersive x-ray spectroscopy (EDS) of these small crystals are consistent with the presence of magnetite (Fig. 3D). While the possibility that other Fe-containing phases are produced in this mutant cannot be completely ruled out, the most parsimonious explanation is that the crystals produced in the absence of the putative protease activity of MamE are indeed magnetite. Furthermore, small crystals were observed within mature sized magnetosome membranes, suggesting that their growth is not restrained by smaller magnetosome membranes (Fig. 3A).
Fig. 2. MamE protease domain is essential for crystal maturation.
A. Crystal size distribution of ΔR9ΔmamE complemented with wildtype, protease inactive (mamEP), and heme-binding deficient mamE (mamEC2). Crystal size is plotted as the percent of the total number of crystals that fall into each 5nm bin. n>500 crystals were measured for each strain from two independent ΔR9ΔmamE/mamE, three independent ΔR9ΔmamE/mamEP, and four independent ΔR9ΔmamE/mamEC2 cultures.
B. Representative TEM images of crystals for each of the three complemented strains. >100 fields were visualized by TEM from >5 independent cultures.
Fig. 3. Electron-dense structures within MamE protease mutant are crystalline.
A. Representative TEM image of a cryo-sectioned ΔR9ΔmamE cell complemented with the MamE protease mutant (mamEP) showing that the small electron-dense 20nm inclusions grow within mature-sized magnetosome membranes. A chain of both empty (white arrows) and inclusion-containing (black arrows) magnetosomes can be observed.
B. HRTEM shows that 20nm electron-dense structures formed when mamEP is expressed in the ΔR9ΔmamE strain, are crystalline, as indicated by the characteristic appearance of the lattice fringes as parallel lines. In the image shown, a measurement of 0.48 nm is consistent with the {111} plane of magnetite. Dotted white lines indicate linear pattern. Scale bar represents 5 nm.
C. HRTEM shows even amorphous electron-dense structures found in this strain are crystalline. Similar to part B, the lattice fringe measurement of 0.25 nm is consistent with the {311} plane of magnetite. Scale bar represents 5 nm.
D. EDX spectra of 20nm inclusions show iron and oxygen peaks consistent with the presence of magnetite. Copper signal is due to copper grids, and phosphorous and carbon peaks from media/cells.
Recently, a second MamE paralog, MamE-like, was annotated outside of the MAI as part of the newly identified magnetosome islet (Rioux et al., 2010). To rule out the possibility that MamE-like can cross-complement MamE protease activity and is responsible for the 20 nm crystals observed in the ΔR9ΔmamE/mamEP strain, the triple mamE deletion strain ΔR9ΔmamEΔmamE-like was generated. The triple deletion strain could be complemented with wildtype mamE to the same extent as the ΔR9ΔmamE and ΔmamE strains. mamEP still supported the growth of small 20 nm crystals in this background (Fig. S3), suggesting that unlike LimE, MamE-like is not capable of cross-complementing protease activity. Together, these data suggest that MamE’s putative protease function is not essential for initiation of biomineralization.
MamE’s protease activity is required for a novel crystal size transition step
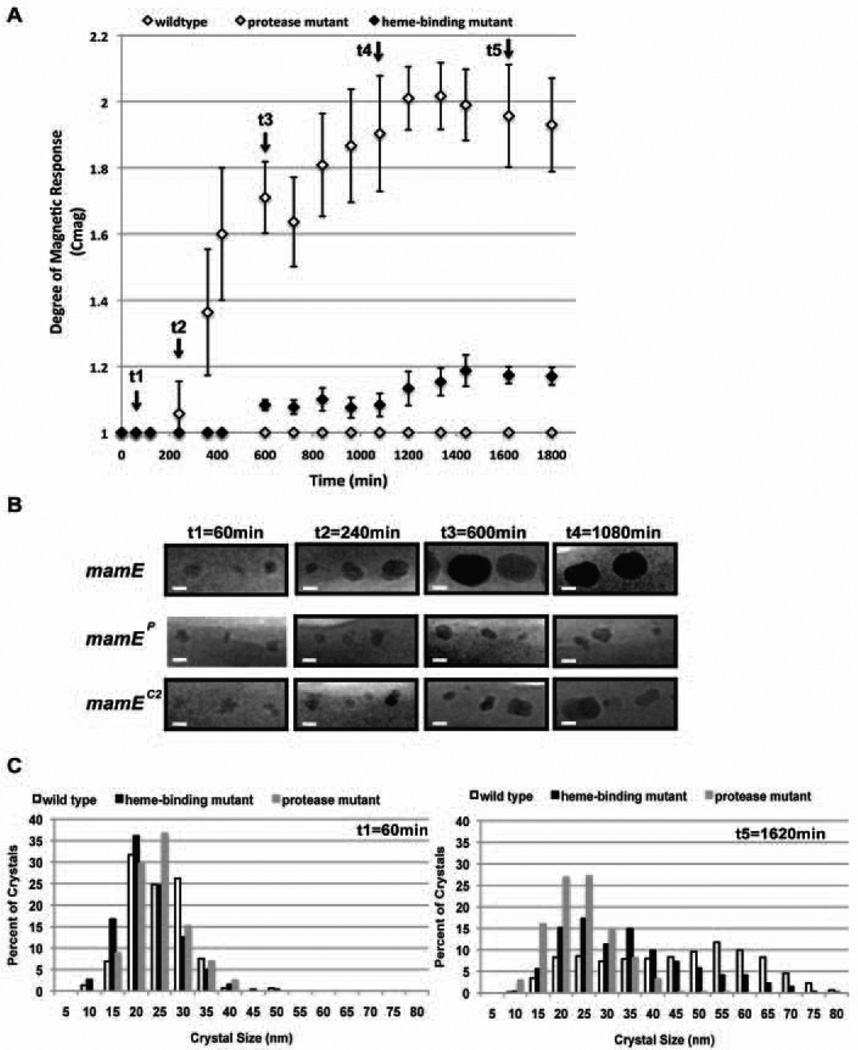
The above results suggest that the putative protease activity of MamE is required for continued growth of magnetite crystals beyond the 16–20 nm size range. The results could also be consistent with a delay in the initiation of biomineralization rather than a change in the kinetics of crystal growth. To distinguish between these two models, we examined the development of the magnetic phenotype and biomineralization of magnetite over time. Briefly, we passaged strains carrying different versions of mamE in the absence of iron until no electron-dense structures were detectable by TEM. Cultures were then moved to iron-containing medium, and their optical densities and Cmag values were monitored over time. Wildtype complemented ΔR9ΔmamE became magnetic four hours post addition of iron and reached its maximum Cmag of ~2 after 1335 minutes. As expected, cells expressing only MamEP do not become magnetic (Fig. 4A).
Fig. 4. MamE protease and heme-binding mutants are not defective in early steps of biomineralization.
A. Time-course of increase in magnetic response of complemented strains. Cells were passaged in the absence of iron and then transferred into iron-containing media to initiate magnetite crystal formation. In contrast to the mamE protease mutant the mamE heme-binding mutant becomes magnetic but is delayed in the onset of its magnetic response as compared to wildtype complemented ΔR9ΔmamE. Error bars represent one standard deviation of three independent cultures. Arrows indicate points at which cells were fixed for TEM imaging (see B) and crystal size determination (see C).
B. Representative TEM images of electron dense inclusions from ΔR9ΔmamE strain complemented with either wildtype mamE, or protease- (mamEP) or heme-binding-deficient (mamEC2) derivatives. At 60 minutes, all three strains produce similar-sized electron-dense inclusions. >150 crystals were visualized and measured at each timepoint.
C. Size distribution of electron dense structures formed by the three strains at early (60 minutes post addition of iron) and late (1620 minutes post addition of iron) time points. Cells were fixed and imaged by TEM at each time point. Crystals were measured and the long axis of the crystal is reported as crystal size. n>150 for 60 minute. n>350 for 1620 minute time point.
To observe the morphological characteristics associated with the biomineralization of magnetite at the early stages, samples obtained at the following points in time were prepared for TEM: (i) before any strain becomes magnetic; (ii) the point at which the wildtype strain becomes magnetic; and (iii) late time points after both strains have been growing in iron for some time. After one hour of growth in iron-containing media, the earliest point in our time-course, both strains had produced electron-dense structures of similar size and shape (Fig. 4B), suggesting that at the time resolution of our experiments, MamEP was capable of initiating biomineralization properly. After three hours, TEM showed large crystals in the ΔR9ΔmamE cells expressing mamE, whereas the same strain carrying mamEP showed only small electron-dense structures (Fig. 4B). In fact in the presence of MamEP, cells were unable to build large crystals, confirming the arrest at the 16–20 nm growth stage seen in the steady-state cultures (Fig 4B and C). Furthermore, the crystal size distributions the ΔR9ΔmamE strain complemented with wildtype mamE or mamEP are remarkably similar at the one hour timepoint (Fig 4C), suggesting that the putative protease function of MamE is not required for initiation of biomineralization but for a potentially unrecognized crystal size transition step. Although the overall trend is similar, for unknown reasons, size distributions measured for these time-courses are slightly shifted towards larger crystals as compared to distributions seen in end point experiments shown in Figure 2. It should also be noted that a small number of crystals (~3%) seem to escape this arrest and attain sizes of greater than 35nm and thus fixed single domain sizes (Fig. 2 and Fig. 4C).
MamE’s putative heme-binding motifs are required for wildtype biomineralization
Unlike most other Deg family proteases, MamE is predicted to have additional functional domains. Specifically, MamE contains two putative CXXCH heme-binding motifs (Fig. 1C), which in c-type cytochromes are known to bind heme covalently via thioether bonds formed through the two conserved cysteines (Bowman & Bren, 2008). To determine whether MamE’s CXXCH motifs are required for magnetosome formation, we used site-directed mutagenesis to change the conserved cysteine residues of these motifs to alanines. This construct, mamEC2, can only partially complement magnetite formation in the ΔR9ΔmamE strain (Fig. 1D), restoring a Cmag of 1.14±0.03 as opposed to 1.84±0.05 when complemented with wildtype mamE. This suggests a role for MamE’s putative heme-binding motifs in crystal formation.
TEM analysis revealed that this mutant is capable of forming large, wildtype-sized crystals. However, complementation with mamEC2 yielded only ~30% of crystals larger than 35 nm, compared to 82% when the ΔR9ΔmamE strain was complemented with wildtype mamE. The crystal size distribution of mamEC2 was shifted to smaller crystal sizes with a bimodal crystal size distribution centered at 16–20 nm and 36–40 nm (Fig. 2). The 16–20 nm peak observed in this strain clearly overlapped with the crystal size distribution of MamEP strain. Additionally, HRTEM and EDX spectroscopy confirmed that the 16–20 nm crystals in the mamEC2-complemented strain are of similar composition to those found in the mamEP-complemented strain (data not shown). Interestingly, in time-course experiments, the strain expressing mamEC2 was always delayed in the onset of its magnetic response as compared to one bearing wildtype mamE (Fig. 4A), even though growth rates of these strains were not significantly different (data not shown). TEM analysis showed that similar to the strain carrying mamEP, the one with mamEC2 does not show obvious defects in the early steps of biomineralization leading to 20 nm crystals. Instead, MamEC2’s decreased number of large magnetite crystals is due to a defect in late steps of biomineralization (Fig. 4B and C).
This suggests that these 20 nm crystals may be an early intermediate in magnetite biomineralization and further supports the hypothesis that MamE is required for the transition from 20 nm crystals to larger crystals that can contribute to a cell’s magnetic response.
MamE is a bifunctional protein with roles in protein localization and biomineralization
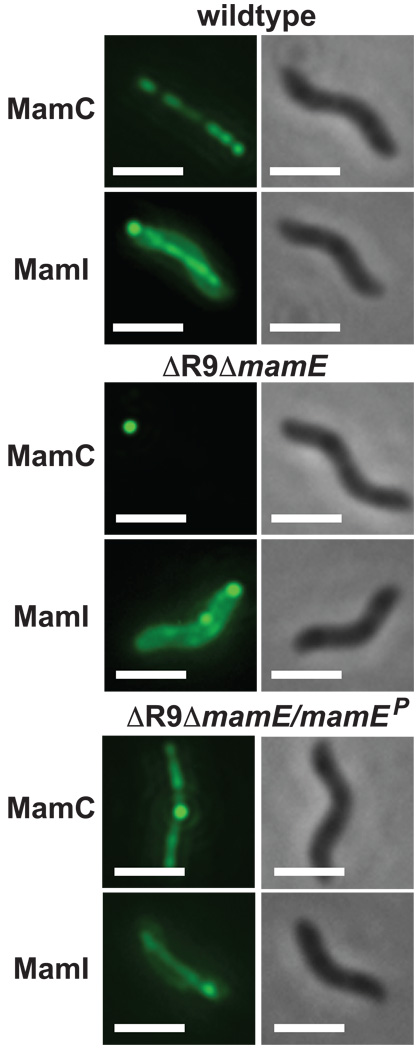
The absence of crystals in the ΔmamE strain had previously been attributed to a defect in magnetosome protein localization as two soluble magnetosome proteins, MamA and MamJ, were mislocalized in this mutant strain (Murat et al., 2010b). To determine whether this defect extends to integral magnetosome membrane proteins, we generated C-terminal GFP fusions to MamC and MamF using a ten-glycine linker, as previously described (Lang & Schuler, 2008). We also assayed localization of GFP-MamI (Murat et al., 2010b). All three proteins form continuous linear structures in wildtype AMB-1 that are reminiscent of the localization of the magnetosome chain. This pattern is disrupted in the ΔR9ΔmamE (Fig. 5 and Fig. S4A) and ΔmamE strains (Fig. S4B). Several localization patterns were observed for the GFP constructs in these deletion backgrounds, ranging from one or multiple foci to evenly distributed membrane localization. This suggests that MamE is required for the proper localization of not only soluble but also membrane-bound magnetosome proteins. It should be noted that in a small percentage of cells, less than 1%, foci were arranged in a linear pattern that could be suggestive of some degree of magnetosome localization (Fig. S4C).
Fig. 5. MamE is a bifunctional protein with a protease-independent role in magnetosome protein localization.
Representative images of localization pattern of MamC-GFP and GFP-MamI and in wildtype AMB-1, in ΔR9ΔmamE and in mamEP complemented ΔR9ΔmamE. These magnetosome proteins form linear structures reminiscent of the localization of the magnetosome chain in wildtype cells that are disrupted in ΔR9ΔmamE. Wildtype localization patterns can be restored with mamEP. >200 cells were imaged. Scale bar represents 1 µm.
It thus seemed likely that MamE’s protease function is required for the proper localization of magnetosome proteins. To test this hypothesis, localization of MamC-GFP and GFP-MamI were assessed in the ΔR9ΔmamE strain complemented by wildtype mamE, mamEP, or mamEC2. Wildtype mamE complemented both the magnetic response (Cmag) and protein localization defects of the ΔR9ΔmamE mutant (Table 1 and Fig 5). However, to our surprise, mamEP, which cannot restore a magnetic response in the ΔR9ΔmamE strain, could complement the protein localization defect. Similar levels of restoration of protein localization were also observed for the strain complemented with mamEC2 (Table 1 and Fig. 5). This suggests that MamE is a bifunctional protein with independent roles in protein localization to the magnetosome and in biomineralization.
Table 1.
Cmag and MamC-GFP and GFP-MamI localization in complemented ΔR9ΔmamE strains. Magnetic response and percent of cells with continuous linear, wildtype-like localization patterns are reported. n is the number of cells scored for GFP localization. Error is reported as one standard deviation representative of variation between different cultures.
| MamC-GFP localization |
Cmag | % of cells with linear localization |
n |
|---|---|---|---|
| ΔR9ΔmamE | 1.0±0 | 1±1 | 259 |
| ΔR9ΔmamE/mamE | 1.52±0.07 | 85±12 | 400 |
| ΔR9ΔmamE/mamEP | 1.0±0 | 75±7 | 464 |
| ΔR9ΔmamE/mamEC2 | 1.10±0.03 | 63±6 | 631 |
|
GFP-MamI localization |
Cmag |
% of cells with linear localization |
n |
| ΔR9ΔmamE | 1.0±0 | 0±0 | 523 |
| ΔR9ΔmamE/mamE | 1.5±0.15 | 82±8 | 1278 |
| ΔR9ΔmamE/mamEP | 1.0±0 | 73±10 | 1061 |
| ΔR9ΔmamE/mamEC2 | 1.07±0.03 | 63±7 | 1018 |
Discussion
To date, most molecular studies of magnetosome formation have focused on identifying magnetosome-associated proteins and determining the phenotypes of deletion or disruption mutants (Grunberg et al., 2004, Tanaka et al., 2006, Fukuda et al., 2006, Komeili et al., 2004, Okuda et al., 1996). These studies have been powerful in providing a list of candidate proteins involved in this intricate process and have laid the foundations for more detailed mechanistic studies. In this work, we used a mutational dissection of the HtrA/DegP family protease MamE to show that this protein acts at two functionally distinct steps of magnetosome formation. First, MamE is required for proper localization of magnetosome proteins. This role does not require MamE protease activity, and proper targeting of magnetosome proteins is not sufficient to produce large magnetic minerals. Since localization of magnetosome proteins does not require MamE protease activity, it could be accomplished through physical interaction of MamE with one or more magnetosome proteins at the magnetosome. Alternatively, it may be mediated by a chaperone-like activity of MamE, which has been attributed to some HtrA/DegP proteins and is separable from their protease activity (Spiess et al., 1999, Rizzitello et al., 2001). Once magnetosome proteins are correctly localized, MamE’s protease activity is required for its second role in magnetosome formation, the maturation of small, 20 nm crystals into larger single-domain crystals with fixed dipole moments. We also show that MamO, a second HtrA/DegP family protease encoded by the MAI, likely has protease activity in vivo. However, further biochemical characterization is required to determine conclusively whether MamE and MamO possess protease activity and if so, what their substrates might be.
Canonical HtrA/DegP proteases consist of a protease domain coupled to one or multiple PDZ domains (Clausen et al., 2002, Kim & Kim, 2005). Strikingly, MamE and MamO have additional functional domains and we show these domains to be important for their function. MamO contains a seven transmembrane spanning domain of unknown function (DUF81). MamO’s paralogue, LimO, is identical to MamO except that it lacks the DUF81 domain (Fig. 1C). Previous proteomic work has suggested that LimO is expressed in wild-type cells (Tanaka et al., 2006). We show that LimO can cross-complement the protease function of MamO when only a protease-inactive version of MamO is expressed in AMB-1. Thus, if LimO is expressed in the ΔmamO strain, as it is the ΔR9ΔmamO/mamOP strain, the non-magnetic phenotype of ΔmamO can be attributed to the lack of a DUF81 domain in LimO. This is consistent with the inability of mamO lacking DUF81 to complement a mamO disruption strain in Magnetospirillum gryphiswaldense MSR-1, a species that does not have a homolog of limO and thus only encodes one copy of mamO (Yang et al., 2010). The function of the DUF81 domain remains mysterious. While it has been suggested to function as an anion transporter in some systems (Weinitschke et al., 2007, Mampel et al., 2004), direct experimental evidence for such an activity is lacking. Alternatively, it is possible that this portion of MamO acts as a localization determinant to bring the protein to the magnetosome.
Similar to MamO, MamE is an unusual HtrA/DegP family protease, in that it also has additional functional domains, two putative CXXCH heme-binding motifs. These motifs are known to bind heme covalently via thioether bonds between the cysteines of the CXXCH motif and the heme vinyl group, and are most commonly associated with c-type cytochromes (Bowman & Bren, 2008). c-type cytochromes have been shown to function in the oxidation and reduction of metals (Paquete & Louro, 2010) and to act as gas sensors (Takayama et al., 2006). MamE could thus act in the reduction and/or oxidation of iron required for magnetite formation. However, MamE cannot be the sole player in this role, since the heme-binding mutant is still capable of biomineralization of mature magnetite crystals. Alternatively, it is possible that MamE’s CXXCH domains are regulatory in nature. Interestingly, two other magnetosome proteins implicated in biomineralization, MamP and MamT, also have CXXCH motifs, raising the possibility that a network of redox activity may be at the center of magnetite biomineralization. Another possible function for the CXXCH motifs is that they could modulate MamE’s protease activity. Time-course experiments showed that although mamEC2 is capable of synthesizing large fixed single-domain magnetite crystals, it is delayed or slowed in this process and forms a significant number of small 20 nm crystals similar to those found in mamEP. This similarity in phenotype to mamEP suggests that MamE’s CXXCH motifs may serve to activate or enhance proteolysis required for crystal size transition.
While it is clear that protease activity of MamE is linked to a specific step of biomineralization, the mechanisms by which this is achieved remain mysterious. We could envision that once a crystal has reached the 20 nm transition point, MamE (i) degrades one or several inhibitors of biomineralization (Fig. 6A), or (ii) proteolytically activates proteins essential for further crystal growth (Fig. 6B). If proteolysis by MamE activates biomineralization factors, then these should be proteins that act in the post-nucleation steps of biomineralization and their deletion phenotypes should be similar to that of the MamE protease mutant. Thus, possible substrates of MamE, based on our current knowledge of biomineralization, could be MamS, MamR, MamT and the proteins encoded by the R2 and R3 genomic regions, which include the mms6 and mamCDFG gene clusters (Arakaki et al., 2003, Scheffel et al., 2008, Murat et al., 2010b). Inhibitors of crystal size transition would have eluded identification by methods currently used to screen biomineralization mutants, since their deletion or disruption would presumably lead to larger magnetite crystals, a phenotype that likely would not have caused a noticeable difference in the cells’ magnetic response. Interestingly, MamE and MamO are not the only examples of serine proteases essential for biomineralization. Enamel proteases have long been known for their role in tooth formation, where they act to remove an organic matrix from nucleated enamel crystallites allowing for growth of the crystallites into mature-sized enamel (Bartlett & Simmer, 1999). Additionally, some serine proteases are capable of precipitating metal oxides in vitro, raising the intriguing possibility that MamE or MamO could play a direct role in the formation of iron oxide crystals (Smith et al., 2009). Thus, identification of MamE and MamO’s substrates may shed light on the potential similarities between the biomineralization pathways of magnetotactic bacteria and those found in other organisms.
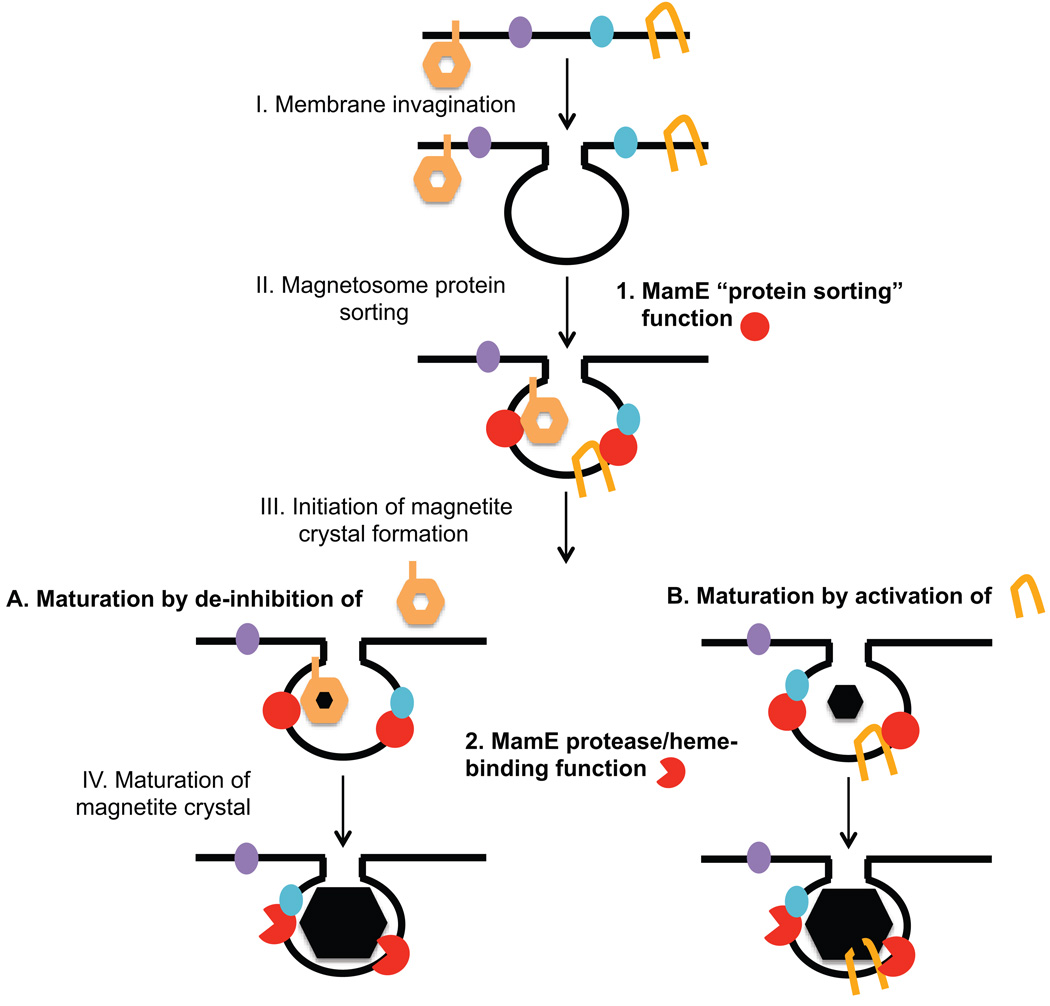
Fig. 6. Model for MamE’s role in magnetosome formation.
After magnetosome membranes are formed MamE is required in a protease independent fashion for the proper localization of at least a subset of magnetosome proteins. This activity is sufficient for the formation of 20nm crystals of magnetite. MamE’s protease function is then required to mature these crystals into large fixed single domain crystals of magnetite. This could be achieved either by (A) proteolytically removing one or several inhibitors of magnetosome formation or an inhibitory matrix (indicated by the orange hexagon), or by (B) proteolytically activating one or several biomineralization promoting proteins. Blue oval: magnetosome membrane protein; purple oval: inner membrane protein; orange symbol: inhibitor or activator of biomineralization; red circle: MamE protease independent function; red pacman: MamE protease-dependent activity. I.–IV.: proposed steps in magnetosome formation.
The work presented here helps define the functional relevance of MamE and MamO in more detail. We speculate that our results may also have uncovered a previously unrecognized “checkpoint” step in magnetite biomineralization where the protease activity of MamE is required for the development of mature magnetite crystals. The transition from small superparamagnetic crystals to larger single domain crystals is an important decision point for the bacterium, one that will trap the cell in a forced biased swim guided by the geomagnetic field, leaving it to use magnetoaerotaxis as its sole mode of exploring the environment. Thus, if MamE’s protease activity could be modulated, cells could arrest biomineralization at the 20 nm stage and be primed for the formation of larger crystals without committing themselves to magnetoaerotaxis. When desirable conditions are reached, biomineralization could be resumed by simply activating MamE protease activity. Such a system would allow cells to align with the earth’s magnetic field lines when favorable without being committed to a forced directional swim under all environmental conditions. In addition, the two-fold increase in the diameter of the crystal represents an approximately eight-fold increase in its volume, meaning that significant resources must be dedicated to build a magnetosome chain after this transition point. MamE’s protease activity could integrate the availability of iron with the need to maximize the production of large single domain crystals. It remains to be determined whether MamE’s protease activity is modulated in vivo, potentially via its putative heme-binding motifs. Regardless of the physiological role of this transitional step in biomineralization, our work has uncovered intriguing possibilities for genetic engineering of MTB where artificial control over MamE protease activity might allow for external control of crystal size and magnetoaerotaxis.
Experimental Procedures
Growth conditions
Cells were grown in defined minimal media (MG medium) supplemented with both Wolfe’s Vitamin Solution and iron (3 mM iron chloride 9 mM malate) at 1/100 (Murat et al., 2010b). Additional iron sources were omitted when preparing Wolfe’s Mineral Solution. For fluorescence microscopy cells, were grown in 10 mL of MG with 20 mL of head space in a microaerobic chamber maintained at 30°C and less than 10% oxygen. To assay complementation and to measure crystal size distributions, cells were grown in a 30°C incubator in sealed Balch tubes containing 10 mL of MG and 20 mL headspace. Balch tubes were flushed briefly with N2 after autoclaving and for 10 minutes after addition of iron and vitamins before cells were added. No additional air was added to the tubes. For time course experiments, cells were grown in Balch tubes, as described above, except that the culture volume was increased to 15ml. For these experiments, all glassware was soaked in oxalic acid for 12 hours before use, and the cells were passaged twice in MG medium containing no iron until the cultures were non-magnetic and no crystals or inclusions were observable by TEM. Solid media plates contained 7g agar per liter medium. Antibiotics were used at the following concentrations: kanamycin at 15 µg/mL in solid media, and at 10 µg/mL in liquid media; 7 µg/mL in liquid when plasmid was integrated on the chromosome; and carbenicillin 2 0µg/mL on solid and liquid media.
Plasmids, Primers and Strains
All primers, plasmids and strains used in this study are listed in Supporting Information tables S1 and S2 and S3.
Time-course experiments
For time-course experiments cells were passaged twice in 10 mL MG medium containing no iron in 20 mL tubes and incubated in a microaerophillic chamber. Cells in exponential phase were then passaged into 15 mL MG medium plus iron malate in Balch tubes and the increase in Cmag was monitored (Komeili et al., 2004). To follow crystal growth, cells were collected by filtration, washed in PBS and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer for TEM analysis. Crystal size was measured by hand using GIMP software and the long axis is reported as crystal size.
Generation of ΔR9ΔmamE, ΔR9ΔmamO and ΔR9ΔmamEΔmamE-like
All deletions were generated using the two-step recombination method previously described (Murat et al., 2010; Komeili et al., 2004). mamE and mamO were deleted in the ΔR9 background using the plasmids pAK241 and pAK243 described in Murat et al., 2010. To delete mamE-like in the ΔR9ΔmamE background, regions flanking the gene were amplified and combined into the deletion plasmid pAK455 by fusion PCR.
Generation of MamC-GFP and MamF-GFP
All PCR except for QuikChange® site directed mutagenesis were carried out using the Promega GoTaq® Green Master Mix. MamC was C-terminally GFP-tagged by fusion PCR. MamC was amplified using a forward primer adding an EcoRI site and a reverse primer deleting the stop codon and adding a BamHI site followed by 10 C-terminal Glycines as a linker. GFP was amplified using a forward primer adding 10 N-terminal Glycines and a reverse primer adding a SpeI site. The MamC-BamHI-10Glycine-GFP construct was then generated by fusion PCR using the MamC forward and GFP reverse primers. This fragment was EcoRI/SpeI cloned into pAK22 to generate pAK452. To confer carbenicillin resistance, the ampicillin resistance gene (bla) placed downstream of the tac promoter was isolated from pAK237 by SpeI digestion and then SpeI cloned into pAK452 to generate pAK453. MamF was amplified adding an N-terminal EcoRI and a C-terminal BamHI site, deleting the stop codon. This fragment was cloned into pAK452 to replace MamC generating pAK454.
Fluorescence microscopy
Cells were imaged in early stationary phase on 1% agarose pads using a Nikon Eclipse 80i microscope. Images were acquired at 1000X magnification using a QImaging® RETIGA 2000R Fast 1394 camera.
Generation of mamO and mamE complementation plasmids
The previously described plasmids pAK263 and pAK250 (Murat et al., 2010b) were used for wildtype complementation of the mamO and mamE deletion strains respectively. To identify mamO and mamE’s putative active site residues, their amino acid sequence was aligned to E. coli DegP, DegQ and DegS using ClustalW2 (Chenna et al., 2003). Putative active site residues were changed to alanines using Stratagene QuikChange® mutagenesis as directed by the manufacturer. MamE’s putative heme-binding motifs were inactivated using the same strategy to change the two cysteines of the CXXCH domains to alanines. The mamO deletion strains were complemented with mamO on a plasmid expressed off of the tac promoter (pAK263) whereas mamE deletion strains were complemented with mamE expressed from its endogenous promoter by integrating mamE into the chromosome (pAK250) (Murat et al., 2010b).
Complementation assays
Cmag measurements were performed with cultures grown in 10ml of MG in sealed 20 mL Balch tubes after two days of growth at 30°C. For mamO complementation, Cmags were measured in the presence of kanamycin. For mamE complementation, Cmags were measured in the absence of any antibiotics. Complementation of protein localization was assayed by growing complemented cells in 10 mL of MG with carbenicillin in 20 mL tubes in a microaerobic chamber.
TEM
For imaging of whole mounts in TEM, a 5 µL drop of the suspended cells was adsorbed onto 200-mesh Cu grid coated with Formvar film. Samples were imaged with an FEI Tecnai 12 TEM equipped with a Gatan Bioscan (1k × 1k) CCD Camera Model 792 at an accelerating voltage of 120 kV.
Cryo-ultramicrotomy and EDS spectroscopy
Cryo-ultramicrotomy was performed as previously described. The sections were imaged with an FEI Tecnai G2 F20 cryo-S/TEM equipped with a Gatan Ultrascan 1000 (4k × 4k) CCD camera and a EDAX Genesis microanalytical system at an accelerating voltage of 200 kV.
Supplementary Material
Acknowledgements
We thank Komeili laboratory members, K. Ryan (also at UC Berkeley) and R.J. Quinlan for their valuable input and discussion both into this work and into preparation of this manuscript. A.Q. was supported by the Winkler Family Foundation. A.K. was additionally funded by the David and Lucille Packard Foundation and the National Institute of Health (5R01GM084122). H.V. acknowledges the Natural Sciences and Engineering Research Council of Canada for financial support.
Contributor Information
Anna Quinlan, Email: anna.wiedmann@gmail.com.
Dorothée Murat, Email: dmurat@berkeley.edu.
Hojatollah Vali, Email: hojatollah.vali@mcgill.ca.
References
- Arakaki A, Masuda F, Amemiya Y, Tanaka T, Matsunaga T. Control of the morphology and size of magnetite particles with peptides mimicking the Mms6 protein from magnetotactic bacteria. J Colloid Interface Sci. 2010;343:65–70. doi: 10.1016/j.jcis.2009.11.043. [DOI] [PubMed] [Google Scholar]
- Arakaki A, Webb J, Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J Biol Chem. 2003;278:8745–8750. doi: 10.1074/jbc.M211729200. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- Bowman SEJ, Bren KL. The chemistry and biochemistry of heme c: functional bases for covalent attachment. Nat Prod Rep. 2008;25:1118–1130. doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler RF, Banerjee SK. Theoretical Single-Domain Grain-Size Range in Magnetite and Titanomagnetite. J Geophys Res. 1975;80:4049–4058. [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. Magnetoaerotaxis in marine coccoid bacteria. Biophys J. 1997;73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst JA. Intracellular compartmentation in planctomycetes. Annu Rev Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Okamura Y, Takeyama H, Matsunaga T. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett. 2006;580:801–812. doi: 10.1016/j.febslet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Grünberg K, Müller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2004;70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K, Wawer C, Tebo BM, Schüler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol. 2005;38:266–274. doi: 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- Komeili A. Molecular mechanisms of magnetosome formation. Annu Rev Biochem. 2007;76:351–366. doi: 10.1146/annurev.biochem.74.082803.133444. [DOI] [PubMed] [Google Scholar]
- Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Schüler D. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl Environ Microbiol. 2008;74:4944–4953. doi: 10.1128/AEM.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampel J, Maier E, Tralau T, Ruff J, Benz R, Cook AM. A novel outer-membrane anion channel (porin) as part of a putatively two-component transport system for 4-toluenesulphonate in Comamonas testosteroni T-2. Biochem J. 2004;383:91–99. doi: 10.1042/BJ20040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D, Byrne M, Komeili A. Cell biology of prokaryotic organelles. Cold Spring Harb Perspect Biol. 2010a;2 doi: 10.1101/cshperspect.a000422. a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A. 2010b;107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y, Denda K, Fukumori Y. Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene. 1996;171:99–102. doi: 10.1016/0378-1119(95)00008-9. [DOI] [PubMed] [Google Scholar]
- Paquete CM, Louro RO. Molecular details of multielectron transfer: the case of multiheme cytochromes from metal respiring organisms. Dalton Trans. 2010;39:4259–4266. doi: 10.1039/b917952f. [DOI] [PubMed] [Google Scholar]
- Richter M, Kube M, Bazylinski DA, Lombardot T, Glockner FO, Reinhardt R, Schüler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JB, Philippe N, Pereira S, Pignol D, Wu LF, Ginet N. A Second Actin-Like MamK Protein in Magnetospirillum magneticum AMB-1 Encoded Outside the Genomic Magnetosome Island. Plos One. 2010;5 doi: 10.1371/journal.pone.0009151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. Journal of Bacteriology. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A, Gardes A, Grünberg K, Wanner G, Schüler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Gietl C, Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 2008;148:1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler D, Frankel RB. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl Microbiol Biotechnol. 1999;52:464–473. doi: 10.1007/s002530051547. [DOI] [PubMed] [Google Scholar]
- Schüler D, Uhl R, Bauerlein E. A Simple Light-Scattering Method to Assay Magnetism in Magnetospirillum Gryphiswaldense. Fems Microbiol Lett. 1995;132:139–145. [Google Scholar]
- Shively JM. Complex intracellular structures in prokaryotes. Berlin: Springer; 2006. p. viii, 379 p. [Google Scholar]
- Smith GP, Baustian KJ, Ackerson CJ, Feldheim DL. Metal oxide formation by serine and cysteine proteases. J Mater Chem. 2009;19:8299–8306. [Google Scholar]
- Smith MJ, Sheehan PE, Perry LL, O'Connor K, Csonka LN, Applegate BM, Whitman LJ. Quantifying the magnetic advantage in magnetotaxis. Biophys J. 2006;91:1098–1107. doi: 10.1529/biophysj.106.085167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- Takayama Y, Kobayashi Y, Yahata N, Saitoh T, Hori H, Ikegami T, Akutsu H. Specific binding of CO to tetraheme cytochrome c(3) Biochemistry-Us. 2006;45:3163–3169. doi: 10.1021/bi051867i. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Okamura Y, Arakaki A, Tanaka T, Takeyama H, Matsunaga T. Origin of magnetosome membrane: Proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics. 2006;6:5234–5247. doi: 10.1002/pmic.200500887. [DOI] [PubMed] [Google Scholar]
- Weinitschke S, Denger K, Cook AM, Smits THM. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C-2 sulfonates. Microbiol-Sgm. 2007;153:3055–3060. doi: 10.1099/mic.0.2007/009845-0. [DOI] [PubMed] [Google Scholar]
- Yang W, Li R, Peng T, Zhang Y, Jiang W, Li Y, Li J. mamO and mamE genes are essential for magnetosome crystal biomineralization in Magnetospirillum gryphiswaldense MSR-1. Res Microbiol. 2010;161:701–705. doi: 10.1016/j.resmic.2010.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.