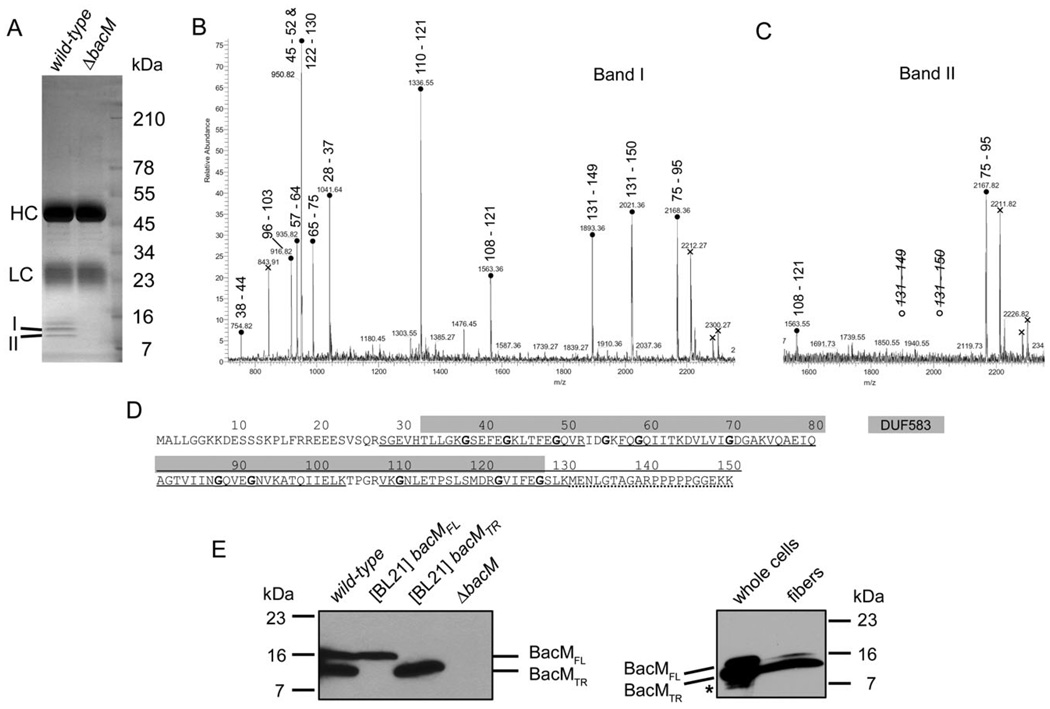
Fig. 3. Processing of BacM.
A. SDS-PAGE gel of the protein A sepharose eluate of an immunoprecipitation with purified anti-BacM antibodies. Used lysates were from strains DK1622 (wild-type) and EH301 (ΔbacM). Band I (13 kDa) and band II (11 kDa) were subjected to MALDI-MS/MS analysis. HC and LC mark the locations of IgG heavy and light chains respectively.
B and C. PMF spectra of tryptic peptides from bands I and II, respectively, with residue numbers attributed to the peaks. Attribution of two different peptides of almost identical mass to the peak at 950.8 Da was confirmed by MS/MS analysis. Crosses indicate peaks originating from trypsin autolysis products. Note the absence of peaks for the C-terminal BacM peptide for band II.
D. BacM amino acid sequence with DUF583 domain (grey box) and sequence coverage of the PMF analysis indicated. Underlined peptides are present in both gel bands, dotted underlining marks presence only in band I. Glycine residues highlighted bold are well-conserved in species from a variety of different taxa (Figs. S1 and S2).
E. Anti-BacM immunoblots showing the different forms of BacM. Left: Cells of wild-type strain DK1622; E. coli BL21-derived strains expressing bacMFL (codon 1–151), or bacMTR (codon 28–151) respectively; and ΔbacM strain EH301. All cells were frozen in liquid nitrogen immediately after harvesting and subsequently dissolved by boiling in Laemmli buffer. BacMFL and BacMTR mark the positions of the full-length, and the truncated version of BacM respectively. Right: whole cellular protein from cells grown on large agar plates. The weak additional bands (asterisk) below the band of BacMTR probably result from proteolysis of BacM due to microscopically confirmed cell lysis on the agar plates prior to harvesting; BacM fibres isolated from these cells in the presence of protease inhibitors produce a strong 13 kDa and a weak 16 kDa band.