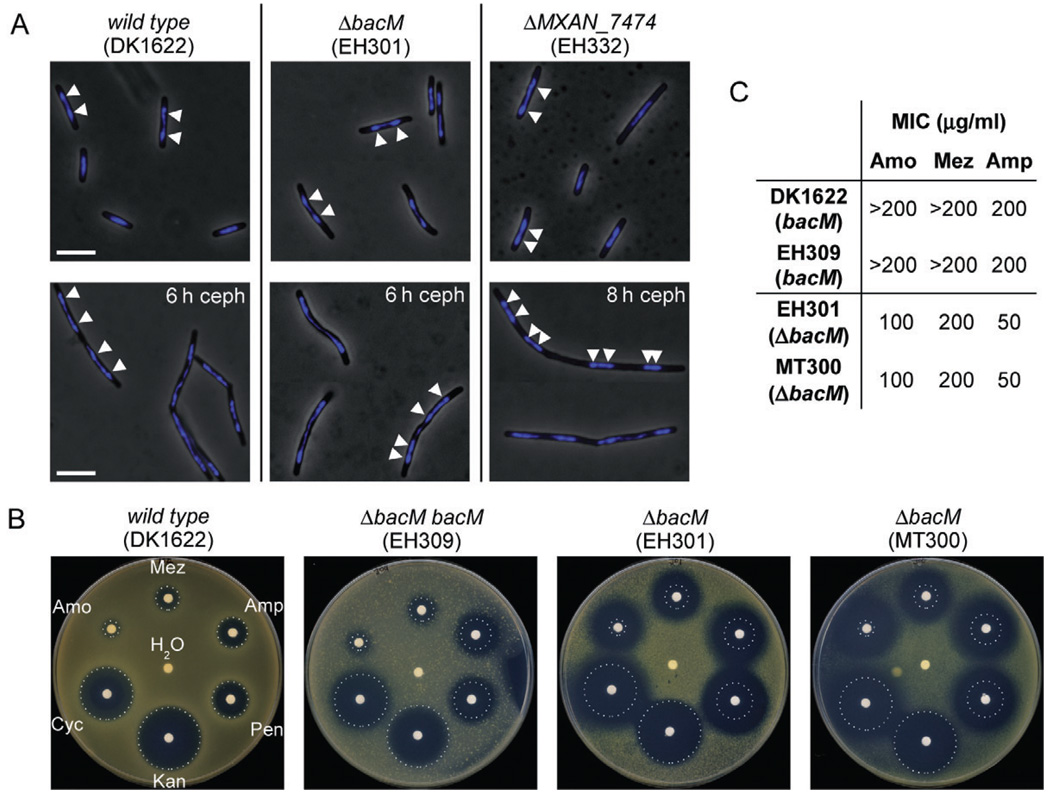
Fig. 7. Chromosome segregation and sensitivity to antibiotics targeting cell wall biosynthesis.
A. Overlays of phase-contrast and DAPI fluorescence micrographs of M. xanthus cells in mid-logarithmic growth phase, or after 6–8 h of growth to OD 0.6–0.8 in medium containing 100 µM cephalexin (ceph). Relevant genotypes are indicated. White bars represent 5 µm. White arrowheads on a selection of cells mark the regions of highest DAPI signal intensity, indicating the centres of the nucleoids.
B. Paper disc diffusion assay comparing antibiotic sensitivities of strains containing or lacking BacM. Antibiotic-containing paper discs were placed on top of CTT agar plates containing cells of the indicated strains. Plates were scanned after 100 h of incubation at 32°C. Sizes of growth inhibition zones of wild-type strain DK1622 are marked by white dotted circles. The discs contained cell wall antibiotics mezlocillin (Mez, 250 µg), ampicillin (Amp, 250 µg), penicillin G (Pen, 100 µg), d-cycloserine (Cyc, 600 µg) and amoxicillin (Amo, 250 mg). Control discs contained water and kanamycin (Kan, 50 µg).
C. Minimal inhibitory concentrations (MIC) of three of the above antibiotics for strains containing or lacking BacM, as determined in 96-well microtitre plate assays (Fig. S11).