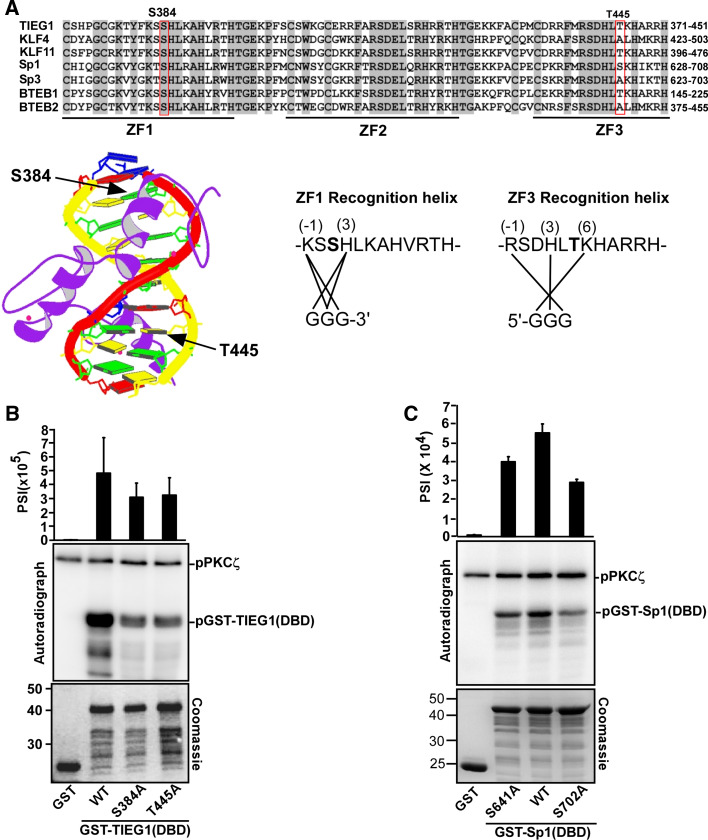
Fig. 3.
PKCζ phosphorylates TIEG1 at Ser 384 located in zinc finger 1 and Thr 445 located in zinc finger 3. a Amino acid sequence alignment of the DBDs of the KLF family proteins. Conserved residues are shaded, and the core zinc finger domains are indicated by thick lines and labeled ZF 1–3. The locations of the phospho-acceptors Ser 384 and Thr 445 residues are boxed. Below and to the left is the structure of the zinc finger protein ZNF268 [57] with the location of the phosphorylation sites in the recognition helixes of zinc fingers 1 and 3 indicated by arrows. Below and to the right are the amino acid sequences of zinc fingers 1 and 3 in TIEG1. The amino acids within the recognition helix that are proposed to contact the DNA bases are indicated by solid lines. The prediction is based on the NMR structure of Sp1 [58]. Ser 384 and Thr 445 are shown in bold. b, c Mutations of TIEG1S384 and TIEG1T445 (b), and Sp1S641 and Sp1S702 (c) to alanine significantly reduce their PKCζ-mediated phosphorylation. Equal amounts of GST and GST-tagged TIEG1 proteins (b), GST and GST-tagged Sp1 proteins (c) were subjected to in vitro phosphorylation assays; detection and quantification as described above. Shown are the quantified phosphorylation levels as bar graphs (upper panels), autoradiographs (middle panels) and Coomassie staining indicating the levels of GST and GST-tagged proteins used in the in vitro kinase assays (lower panels). Each bar in b and c represents the mean (± SD) of four independent experiments