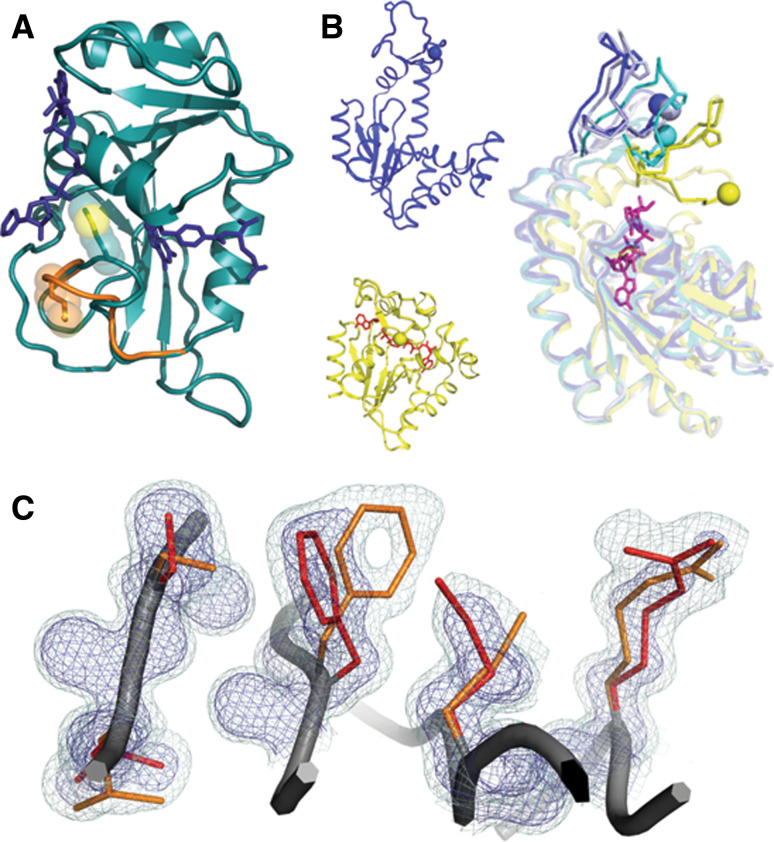
Fig. 1.
Catalytically important motions detected by NMR relaxation experiments can arise through different structural mechanisms. a Polysterism revealed by comparison of NMR chemical shifts with reference to independent crystal structures in DHFR (1RC4 and 1RB2). The Met20 loop of DHFR samples a minor conformational substate (shown in orange) when bound to the substrates NADP+ and THF. The minor state chemical shift is correlated with the chemical shift of the major conformational substate for the immediately preceding state of the catalytic cycle. b Independent crystal structures of AdK in the apo/open state (blue 2RH5) and nucleotide-bound/closed state (yellow, with nucleotide in red 2RGX) reveal that subdomain lid closure underlies the NMR relaxation signal. In an orthogonal view, independent chains from one crystallographic asymmetric unit (blue, light blue, and cyan 2RH5) reveal that the transition path towards the closed state (yellow) is populated even when no nucleotide is bound. Residues on mobile loops are shown in stick and sphere to provide a visual landmark of the polysterism. c In the room-temperature structure of CypA (3K0N), interconverting side-chain conformations give rise to an NMR relaxation dispersion signal. The minor (orange) and major (red) conformational substates extending from the core (left) to the active site Arg55 (right) are shown within electron density mesh. Examining the electron density at low levels (light blue 0.3σ) below normal contour (dark blue 1σ) reveals the minor conformational substate