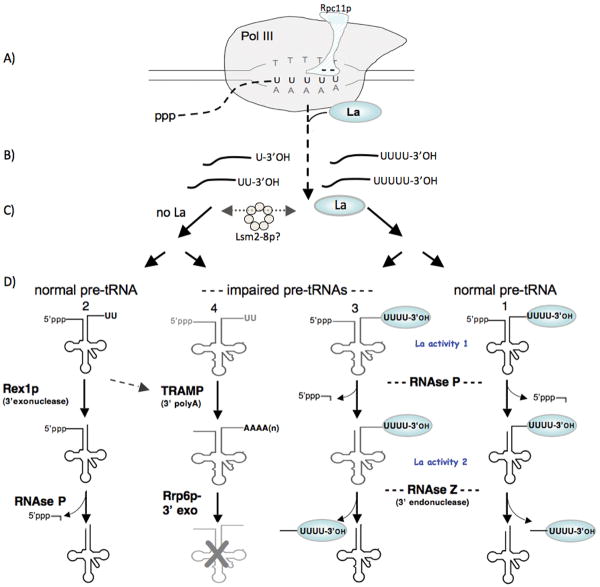
Figure 1.
Alternative pathways for pre-tRNA processing begin during transcription termination by RNA polymerase III. This figure incorporates protein nomenclature from multiple organisms (see text). A) Pol III pauses at the tRNA gene terminator. According to structure modeling the integral pol III subunit, Rpc11p, contains a N-terminal domain that resides on the surface of pol III and an acidic hairpin that enters through a pore to the catalytic center. The acidic tip of the Rpc11p hairpin (designated “- -”) mediates RNA 3′ cleavage of nascent pre-tRNA transcripts during pausing by pol III. B) Nascent pre-tRNA transcripts with different length 3′ oligo(U) termini are released from pol III and sorted into those that bind La and those that do not, depicted on the right and left sides of the vertical dashed arrow respectively. C) Lsm proteins (presumably Lsm2p-8p) can affect the degree to which nascent pre-tRNAs are associated with La and may function at this point (see text). Pre-tRNA transcripts with sufficient 3′ oligo(U) length maintain stable association with La protein. D) Pre-tRNAs move along alternative pathways, numbered 1-4, depicted for normal pre-tRNAs (pathways 1 & 2) and structurally-impaired pre-tRNAs (pathways 3 & 4), distinguished by 3′ endonucleolytic (pathways 1 & 3), or 3′ exonucleolytic (pathways 2 & 4), as well as nuclear surveillance involving TRAMP and Rrp6p (pathway 4). La protein gains early dominance over the pre-tRNAs with sufficient 3′ oligo(U) length (see text). The exact point in this scheme at which the RNA chaperone-like activity of La (La activity 2) appears to function is unclear from current data but occurs sometime before RNase Z separates the trailer from the tRNA body.