Figure 1.
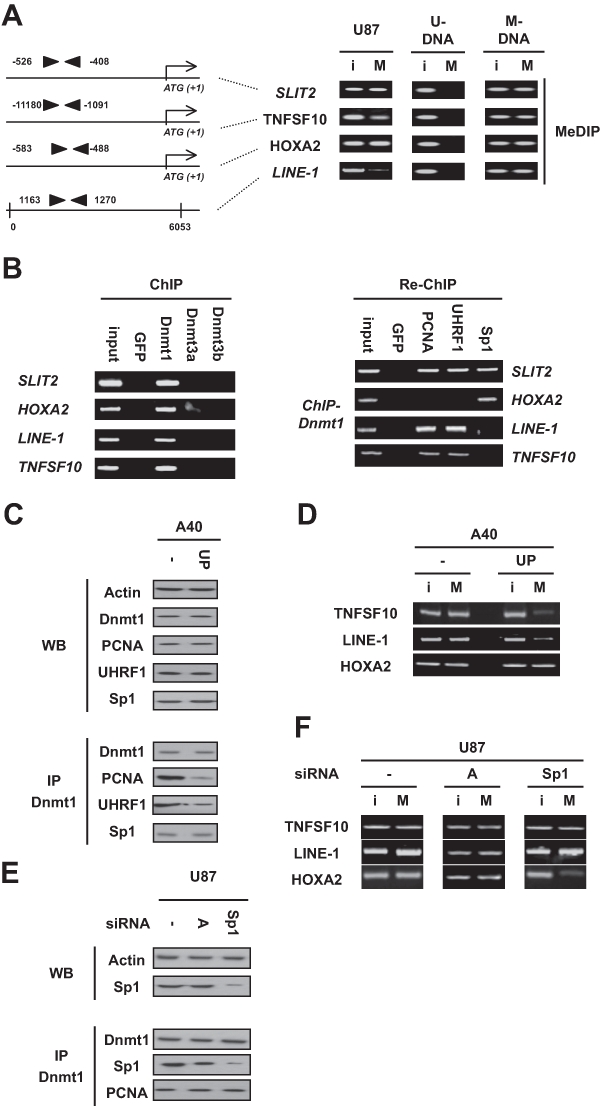
Dnmt1/Sp1 or Dnmt1-PCNA-UHRF1 acts as a single player to maintain the methylation of the −1180/−1091 region of TNFSF10 and the 1163/1270 region of LINE-1 or the −583/−488 region of HOXA2. (A) The methylation status of the SLIT2 (ENSG00000145147), TNFSF10 (ENSG00000121858), and HOXA2 (ENSG00000105996) genes was determined by methylation-specific PCR (MSP). U = unmethylated; M = methylated. The methylation status of LINE-1 (L19092.1) was determined by the methylated DNA immunoprecipitated method (MeDIP). I = input, i.e., no immunoprecipitated DNA; M = MeDIP, i.e., immunoprecipitated DNA. Black triangles represent primers used in MeDIP. U-DNA = CpGenome Universal unmethylated DNA set and M-DNA/CpGenome Universal methylated DNA set (Chemicon, Millipore, Molsheim, France). (B) In U87 cells, chromatin immunoprecipitation (ChIP) experiments were performed to determine the presence of Dnmt1, Dnmt3a, or Dnmt3b on indicated genes. Re-chromatin immunoprecipitation (Re-ChIP) experiments were performed to determine the presence of the considered proteins on indicated genes. (C, D) Impact of the disruption of the Dnmt1-PCNA-UHRF1 interaction on the methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1. The disruption of the Dnmt1-PCNA-UHRF1 interactions via the nucleofection of the Astro40 cells with the pUP plasmid (UP) or plasmid control was monitored by the immunoprecipitation of the Dnmt1 (IP-Dnmt1). Western blot illustrates the expression of Dnmt1, PCNA, UHRF1, and Sp1 in indicated cells. The methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1 was assessed by the methylated DNA immunoprecipitated method (MeDIP). I = input, i.e., no immunoprecipitated DNA; M = MeDIP, i.e., immunoprecipitated DNA. (E, F) Impact of the disruption of the Dnmt1/Sp1 interaction on the methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1. The disruption of the Dnmt1/Sp1 interactions via the Sp1 down-regulation in Astro40 cells by siRNA-Sp1 (sc#29487; Tebu-Bio, Le-Perray-en-Yvelines, France) was monitored by the immunoprecipitation of Dnmt1 (IP-Dnmt1). siRNA-A (sc#37007; Tebu-Bio) is used as control. The methylation status of SLIT2, TNFSF10, HOXA2, and LINE-1 was assessed by the methylated DNA immunoprecipitated method (MeDIP). I = input, i.e., no immunoprecipitated DNA; M = MeDIP, i.e., immunoprecipitated DNA.
