Figure 2.
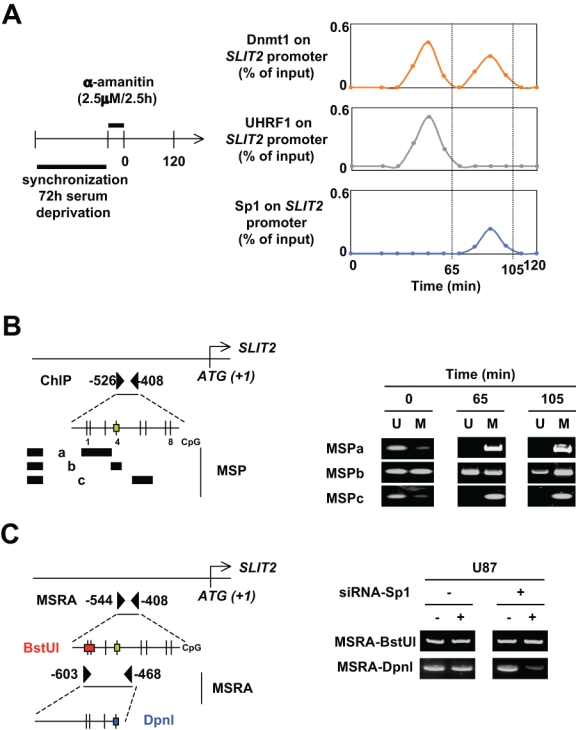
Description of the cooperation between Dnmt1/Sp1 and Dnmt1-PCNA-UHRF1 to maintain the DNA methylation. (A) The kinetic of ChIP experiments was realized at an indicated time after cell (U87) synchronization (72 hours of serum deprivation) and α-amanitin treatment. ChIP was performed with the EZ ChIP kit (Millipore, Molsheim, France) and with indicated antibodies. The amount of immunoprecipitated promoter was quantified by semiquantitative PCR and normalized to input. (B) Identification by methylation-specific PCR (MSP) of the methylation status of CG dinucleotides localized into the −526/−408 region of the SLIT2 gene in U87 cells. MSPa was designed to investigate the methylation status of CG dinucleotides #1, #2, and #3; MSPb was designed to investigate the methylation status of CG dinucleotide #4; and MSPc was designed to investigate the methylation status of CG dinucleotides #5 and #6. The green box indicates the presence of Sp1 binding site according to the use of the Patch program. U = unmethylated; M = methylated. Black boxes represent primers. (C) Impact of siRNA-induced decrease expression of Sp1 on the methylation status of specific CG dinucloetides of the SLIT2 genes. Methylation status of specific CG dicnuleotides was determined by methylation sensitive restriction assay (MSRA). DNA methylation blocks the cleavage of BstUI and DpnI when the CGCG and GATCg sequences are methylated, respectively. Thus, when DNA is initially unmethylated, no or weak DNA is amplified by PCR. Black triangles represent primers.
