Figure 1.
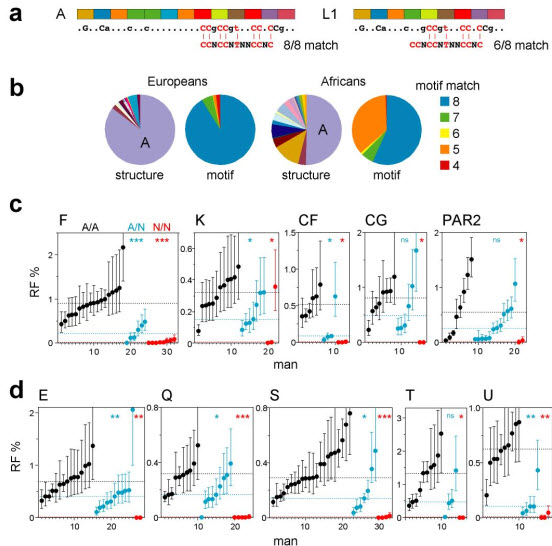
PRDM9 ZnF variants and crossover hot-spot activity in sperm. (a) Examples of tandem repeats encoding the ZnF array, with variant repeat units coloured differently. The predicted DNA binding sequence is shown below, with dots indicating weakly and uppercase the most strongly predicted bases, and aligned with the hot-spot motif CCNCCNTNNCCNC (ref. 4). The binding sequence for allele A matches all 8 specified bases in the motif, while allele L1 matches at best only 6 of the 8 bases. (b) PRDM9 ZnF array diversity in Europeans and Africans, with alleles classified either by structure or by the strength of the predicted match with the motif (details of alleles provided in Supplementary Fig. 1). (c) Variation between men in sperm crossover activity in hot spots, named above each panel, containing a central hot-spot motif. Different sets of men informative for SNPs required for crossover detection were analysed at each hot spot (Supplementary Table 1). Men carrying two PRDM9 A alleles (A/A, black), one A allele (A/N, blue) or two non-A alleles (N/N, red) were grouped separately in ascending order. Confidence intervals for each estimate of RF are shown, and median RFs within each group are indicated by dotted lines. Mann-Whitney test results for the significance of differences between the A/A group and the A/N or N/N groups are given at top right (ns P > 0.05; * P < 0.05; ** P < 0.01; *** P < 0.001). (d) Corresponding analyses of hot spots lacking an obvious hot-spot motif (Supplementary Fig. 2).
