Figure 2.
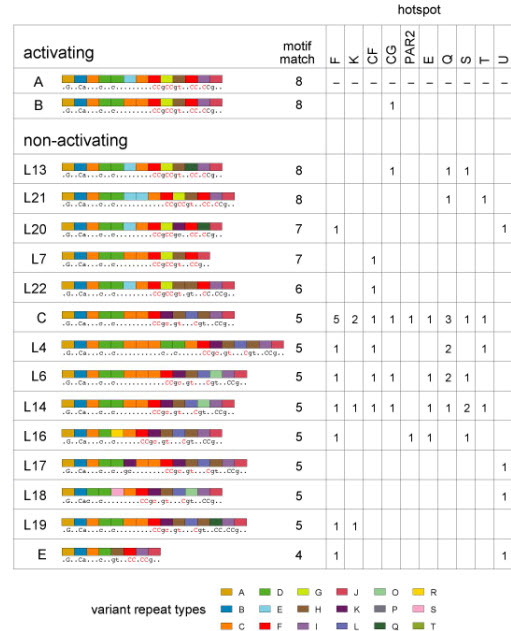
Activating and non-activating PRDM9 alleles. Non-activating alleles at each hot spot were defined as alleles present in N/N men who showed <5% of the median RF seen in A/A men. Allele structures are coded as in Supplementary Fig. 1, with predicted DNA binding sequences and best motif matches shown as in Fig. 1. Data for each hot spot give the number of specific N alleles detected in suppressed men; for example, 5 such men typed at hot-spot F carried the C allele. Evidence that the B allele is active is based on a B/L6 heterozygote assayable only at hot-spot CG who showed crossovers at 40% of the median frequency seen in A/A homozygotes. Since allele L6 is a non-activator at CG, this implies that B is similar in activity to A.
