Abstract
There is increasing evidence that glial cell line-derived neurotrophic factor (GDNF) plays a role as a limiting, striatal target-derived neurotrophic factor for dopamine neurons of the substantia nigra pars compacta (SNpc) by regulating the magnitude of the first phase of postnatal natural cell death which occurs in these neurons. While it has been shown that GDNF mRNA is relatively abundant in postnatal striatum, the cellular basis of its expression has been unknown. We therefore used nonradioactive in situ hybridization and immunohistochemistry to examine the cellular basis of GDNF mRNA and protein expression, respectively, in postnatal striatum and related structures. We found that GDNF mRNA is expressed within medium-sized striatal neurons. Expression in glia was not observed. At the protein level, regionally, GDNF expression in striatum was observed in striosomal patches, as previously described. At a cellular level a few neurons were observed, but they do not account for the striosomal pattern. This pattern is predominantly due to GDNF-positive neuropil. Some of this neuropil arises from tyrosine hydroxylase-positive nigro-striatal dopaminergic afferents. Astrocytic processes do not appear to contribute to the striosomal pattern. GDNF-positive fibers are identified not only within intrinsic striatal neuropil, but also in fibers within the major striatal efferent targets: the globus pallidus, the entopeduncular nucleus, and the SN pars reticulata. We conclude that during normal postnatal development, medium-sized neurons are the principal source of GDNF within the striatum.
Indexing terms: natural cell death, dopamine, substantia nigra, striosome, globus pallidus, GDNF
Glial cell line-derived neurotrophic factor (GDNF) was originally identified based on its ability to support the development of embryonic dopamine neurons in culture (Lin et al., 1993). Since that time it has been demonstrated to both protect and restore dopamine neurons in a variety of lesion models in different species (Tomac et al., 1995a; Bowenkamp et al., 1995; Sauer et al., 1995; Gash et al., 1996; Choi-Lundberg et al., 1997; Kordower et al., 2000). Based on these numerous studies in preclinical models, GDNF is now being evaluated for the treatment of human Parkinson’s disease (Gill et al., 2003).
In spite of its clear effects on the neurobiology of mesencephalic dopamine neurons, very little has been known of the normal physiologic role of GDNF in supporting or maintaining these neurons in living brain. Early work on patterns of developmental expression of GDNF mRNA suggested that it may serve as a target-derived factor for mesencephalic dopamine neurons, because it is highly expressed in striatum in the early postnatal period (Schaar et al., 1993; Stromberg et al., 1993; Blum and Weickert, 1995; Choi-Lundberg and Bohn, 1995). It is during the first two postnatal weeks that dopamine neurons of the substantia nigra (SN) undergo a developmental natural cell death event (Janec and Burke, 1993; Oo and Burke, 1997; Jackson-Lewis et al., 2000) which is biphasic, with peaks at postnatal day (PND) 2 and 14. We have presented evidence that striatal target-derived GDNF may play a physiologic role in regulating the magnitude of the first phase of natural cell death among dopamine neurons on PND2. In an early postnatal primary culture model of mesencephalic dopamine neurons, GDNF is unique in its ability to support viability by suppressing apoptosis (Burke et al., 1998). We have shown in vivo that intrastriatal injection of GDNF during the first phase of natural cell death suppresses apoptosis, whereas injection of neutralizing antibodies augments it (Oo et al., 2003). More recently, we have shown that regionally selective overexpression of GDNF in the striatum throughout development in a double transgenic mouse model induces an increased surviving number of SN dopamine neurons following the first phase of cell death (Kholodilov et al., 2004).
In spite of this growing evidence that striatal GDNF plays a physiologic role in regulating the developmental natural cell death event of SN dopamine neurons, there is a paucity of information about the cellular basis of striatal GDNF expression during development. At the mRNA level, a large majority of developmental studies have been performed on tissue homogenates. The few in situ hybridization studies which have been performed were either not conducted at a cellular level (Stromberg et al., 1993; Golden et al., 1999), or were performed by assessment of autoradiographic silver grain deposition (Trupp et al., 1997), which is not conducive to a high-quality morphologic assessment of the underlying cellular phenotype. Although it is generally assumed that GDNF mRNA is likely to be more highly expressed in striatum than SN, in keeping with its proposed role as a target-derived factor for dopamine neurons, there has never been a direct comparison of their GDNF mRNA levels during development. At the protein level, there is likewise little information available about the cellular basis of expression. While striatal expression of GDNF proteins has been demonstrated at a regional level during development (Lopez-Martin et al., 1999), it has not been examined at the cellular level. We therefore explore these issues by use of Northern analysis, a nonradioisotopic in situ hybridization technique for the demonstration of GDNF mRNA, and by immunohistochemistry for GDNF protein in conjunction with double-labeling for other cellular antigens.
MATERIALS AND METHODS
Northern analysis
Animals
Timed pregnant Sprague-Dawley rats (Charles River Labs, Wilmington, MA) were used to obtain striatum and SN at PND2 and 14. The day of delivery was defined as PND1. SN and striatum tissue were obtained by microdissection as previously described (El-Khodor et al., 2001). The striatum was obtained with a 2-mm (PND2) or 3-mm (PND14) diameter punch. For SN, the midbrain ventral to a horizontal cut below the cerebral aqueduct was obtained. The tissues were then frozen on dry ice and stored at −80°C until RNA isolation. This protocol has been approved by the Institutional Animal Care and Use Committee at Columbia-Presbyterian Medical Center.
Northern analysis
Northern analysis was performed as previously described (El-Khodor et al., 2001). Briefly, RNA was isolated using a Qiagen RNAeasy Mini kit. The RNA concentration of each sample was determined by measuring absorption at 260 nm on a GenQuant spectrophotometer (Amersham Pharmacia Biotech, Piscataway, NJ). Twenty micrograms of RNA was electrophoresed in 1.4% agarose-formaldehyde gel and transferred onto an Immobilon membrane (Millipore, Bedford, MA). To create a probe for Northern analysis, oligonucleotide primers for rat GDNF mRNA were designed based on the published sequence (GenBank Accession no. for GDNF, L15305). Primers were synthesized by GeneLink (Tarrytown, NY). The forward primer was 5’-ACGGGACTCTAAGATGAAGTTA-3’, and the reverse primer was 5’-GTCAGATACATCCACACCGT-3’. One microgram of the striatum RNA was used for cDNA synthesis with a Promega (Madison, WI) reverse transcription system using the recommended conditions. Two microliters of cDNA were then used for PCR, which was carried out in 20 µl of Taq DNA polymerase (Roche, Indianapolis, IN). PCR was performed in an Mastercycler (Eppendorf Scientific, Hamburg, Germany) as follows: 95°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 58°C for 20 seconds, and 72°C for 30 seconds, and then a final elongation step at 72°C for 7 min. PCR products were cloned in pGEM-T vector (Promega), and its sequence confirmed with an ABI PRISM Big Dye Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) followed by analysis on an ABI PRISM 310 DNA Sequencer (Foster City, CA). Plasmid was linearized by cutting with Nco I and used for generation of a 32P-labeled antisense riboprobe with a transcription system using SP6 RNA polymerase. The hybridization was performed overnight at 68°C in Ultrahyb buffer (pH 7.5) from Ambion (Austin, TX). The membrane was then dried and exposed to phosphor imager cassettes, scanned, and analyzed by Image Quant software (Molecular Dynamics, Indianapolis, IN). Radioactive bands were expressed as a relative percentage of the radioactivity within the striatum sample at PND2 on each membrane. Two animals representing each region and each time point were represented on each blot. Four independent hybridizations were performed for n = 8 for each region at PND2 and 14. We used quantitative analysis of total RNA and inspection of ethidium bromide-stained gels to ensure equal loading of lanes. We have previously shown that GAPDH mRNA expression, often used as an indicator of total mRNA loading, is developmentally regulated (El-Khodor et al., 2001), so that normalization by GAPDH mRNA levels results in a spurious, decreased apparent level of expression of the mRNA of interest between postnatal weeks 2–4. Similarly, α- and β-tubulin (Bhattacharya et al., 1987) and β-actin (Lazarini et al., 1991; Micheva et al., 1998) mRNA are developmentally regulated. For these reasons, we did not normalize mRNA determinations by the mRNA for these genes in these developmental studies.
Nonradioactive in situ hybridization (NRISH)
Rats (n = 4) at PND5–7 were anesthetized deeply with halothane and then perfused intracardially with 0.9% saline by gravity for 5 minutes, followed by 4% paraformaldehyde (PF) in 0.1 M phosphate buffer (PB) (pH 7.1) for 10 minutes at 4°C. The brain was carefully removed and the SN or striatum was blocked. Brains were postfixed in 4% PF/0.1 M PB for 3 hours at 4°C and cryoprotected in 20% sucrose in 0.1 M PB for 24 hours prior to sectioning. Then brains were rapidly frozen by immersion in 2-methylbutane on dry ice and sectioned on a cryostat at 20 µm. The sections were serially collected into wells containing 0.1 M phosphate-buffered saline (PBS) (pH 7.1), mounted onto Superfrost slides (Fisher Scientific, Hampton, NH) slides, and dried for 1 hour at 40°C. For hybridization, the sections were washed in PBS and then acetylated in triethanolamine / acetic anhydride solution for 10 minutes at room temperature (RT). Following a wash in PBS, sections were prehybridized for 2 hours at RT as previously described (Burke et al., 1994). Digoxigenin-11-UTP riboprobes were prepared as per the manufacturer’s instructions (Roche Diagnostics, Penzberg, Germany). The size and integrity of the labeled probe were confirmed by gel electrophoresis. Hybridization was performed at 68°C overnight in a humidified chamber. After hybridization, sections were washed in 5 × saline sodium citrate (SSC) (for 10 minutes) and 0.2×SSC (3× for 30 minutes) at 68°C, then treated with 0.1 M Tris / 0.15 M NaCl / 10% sheep serum for 1 hour at RT and incubated overnight with an anti-digoxigenin antibody at 1:5,000 (Roche Diagnostics, Mannheim, Germany). Following a wash in 0.1 M Tris / 0.15 M NaCl, sections were incubated with a solution containing nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate (Promega) in a darkened humidified chamber overnight. Sections were then washed in Tris/EDTA (pH 8.0) and coverslipped with aqueous mounting medium (Dako).
GDNF immunostaining
GDNF immunoperoxidase staining
Sprague-Dawley rat pups were used for study at PND5 (during the first phase of natural cell death) and at PND14 (during the second phase). Rat pups were deeply anesthetized and perfused and their brains removed. For regional analysis of immunostaining, brains were fixed and cryoprotected as described for NRISH. Coronal sections of the SN, striatum, globus pallidus (GP), and entopeduncular nucleus (EP) were cut at 30 µm on a cryostat and processed free-floating. Horizontal sections were cut at 40 µm. After a wash in 0.1 M PBS, sections were treated with 0.1 M PBS / 5% horse serum, and then 0.1 M PBS / 5% horse serum / 0.1% Triton for 30 minutes. Sections were then incubated with goat anti-GDNF (AF-212-NA) (R&D Systems, Minneapolis, MN) at 1:500 for 24 hours at RT with shaking. After a wash in PBS / 5% horse serum, sections were incubated in biotinylated-horse-antigoat (BA-9500) (Vector Laboratories, Burlingame, CA) at 1:200 for 2 hours at RT. After a wash, sections were then treated with avidin-biotinylated horseradish peroxidase complexes (ABC, Vector Laboratories) for 1 hour at RT. After washes in PBS (2× for 5 minutes each), sections were incubated in a solution of diaminobenzidine (50 mg in 100 ml Tris buffer at pH 7.6) containing glucose oxidase, ammonium chloride, and D(+) glucose to generate H2O2. Following three 15-minute washes in Tris buffer, pH 7.6, sections were then mounted on gelatin-coated slides and left to dry overnight at RT. Sections were then dehydrated through alcohols, cleared in xylene, and coverslipped under Permount. In order to stain cell bodies for GDNF, the brain was embedded in 6% agar and 100-µm sections were cut using a Vibratome. Sections were washed with 5% normal horse serum followed by PBS / 5% normal horse serum / 0.1% Triton for 1 hour at 4°C. After additional washes in PBS, sections were incubated in anti-GDNF antiserum at 1:250 overnight at room temperature. The sections were then processed as described for regional immunostaining. The primary antibody used in this analysis (R&D Systems, AF-212-NA) has previously been validated as monospecific for the demonstration of both rat and human GDNF in immunohistochemical studies by Western analysis and preabsorption controls (Pina et al., 2002; Kirik et al., 2000; Georgievska et al., 2002; Kordower et al., 2000). In addition, we have shown that this antibody does not crossreact with other members of the GDNF family of ligands (Oo et al., 2003).
GDNF and NeuN immunofluorescence double-labeling
Rat pups aged PND5–7 were perfused and their brains removed, postfixed, frozen, and sectioned as described for GDNF peroxidase labeling. Sections were washed in PBS, treated with PBS / 5% horse serum / 5% donkey serum, and then with 0.1% Triton in PBS / 5% horse serum / 5% donkey serum. The sections were then incubated with goat anti-GDNF (1:500) and a mouse anti-NeuN monoclonal antibody (1:100) (MAB-377) (Chemicon, Temecula, CA) at RT for 24 hours. After a wash in PBS, sections were then incubated in donkey antigoat-AlexaFluor594 (1:250) (Molecular Probes) and horse antimouse-FITC (1:75) (Vector Laboratories) at RT for 2 hours. Sections were then washed in PBS, mounted onto gelatin-coated slides, dried, and coverslipped with DAKO anti-fade medium. Fluorescence by the appropriate fluoro-phore was completely eliminated by the exclusion of the primary antibody antigen. Sections were analyzed with appropriate filters either by epifluorescence on a Nikon Eclipse E800 microscope or by confocal microscopy on a Zeiss LSM 510 META system.
GDNF and GFAP double-labeling
Sections were processed as described above, except the primary antibody used for GFAP was a mouse monoclonal cocktail (1:100) (RDI-GFAPabm-CK) (Research Diagnostics, Flanders, NJ).
GDNF and tyrosine hydroxylase (TH) double-labeling
Sections were processed as described above, except the primary antibody used for TH was a mouse monoclonal (1:40) (MAB 5280) (Chemicon).
Digital photography
Photomicrographic images were captured on a Nikon Eclipse 800 microscope with a Spot 2 digital camera and imported into Adobe PhotoShop (San Jose, CA). Images were modified in brightness and contrast to optimize photographic representation of images observed under the microscope. There was no further digital modification of the images.
Statistical analysis
The differences in levels of GDNF mRNA expression at PND2 and 14 were examined for significance by t-test using the SigmaStat program (Systat, Point Richmond, CA).
RESULTS
GDNF mRNA expression in striatum and SN
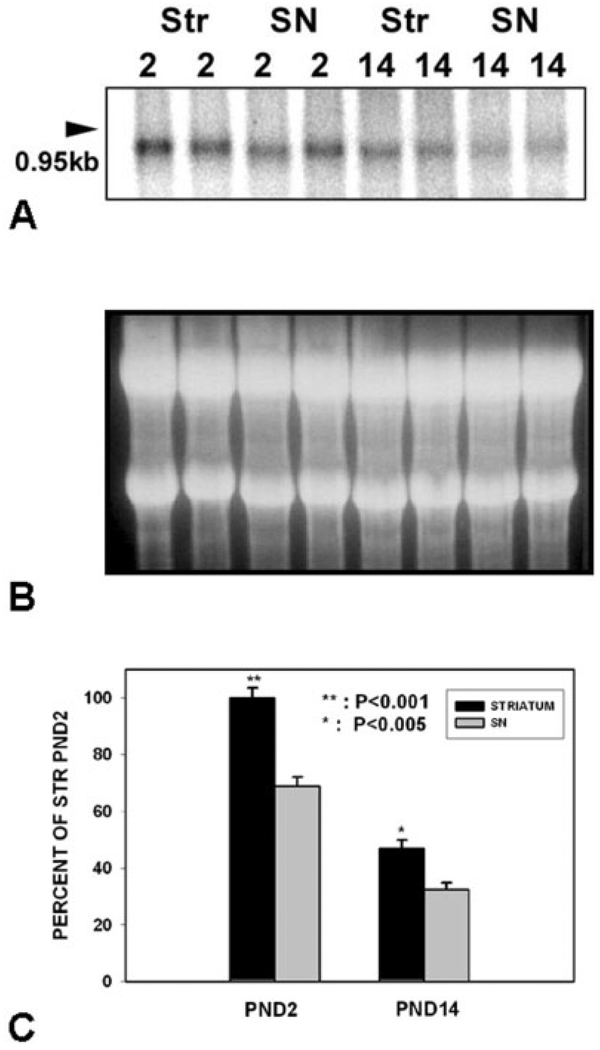
To determine the relative abundance of GDNF mRNA in SN and striatum during the postnatal peaks of natural cell death in dopamine neurons, we performed Northern analysis on these regions at PND2 and 14 (Fig. 1). Northern analysis during this developmental period revealed a single band of ~ 900 bp in both striatum and SN (Fig. 1A). A similar size mRNA species has been reported previously for rat GNDF mRNA (Abe and Hayashi, 1997). Northern analysis revealed that striatal GDNF mRNA levels are higher on PND2 than PND14, in keeping with our previous developmental analysis (Cho et al., 2003). In keeping with a role as a possible target-derived neurotrophic factor, higher levels of GDNF mRNA were expressed in striatum than SN (Fig. 1C). Nevertheless, substantial levels of GDNF mRNA were expressed in SN.
Fig. 1.
Relative levels of GDNF mRNA in striatum and SN at the two phases of NCD in SN. A: Representative Northern analysis of GDNF mRNA expression in striatum and SN at PND2 (n = 2 for each region) and PND14 (n = 2 for each region). B: An ethidium bromide stain of the gel demonstrates equal loading of the lanes. C: Quantitative analysis of GDNF mRNA expression in striatum and SN at the two phases of NCD in SN (n = 8 for each condition). Note that mRNA levels were relatively greater in striatum than SN during both phases of NCD, but particularly at PND2 (P < 0.001 at PND2, P < 0.005 at PND14).
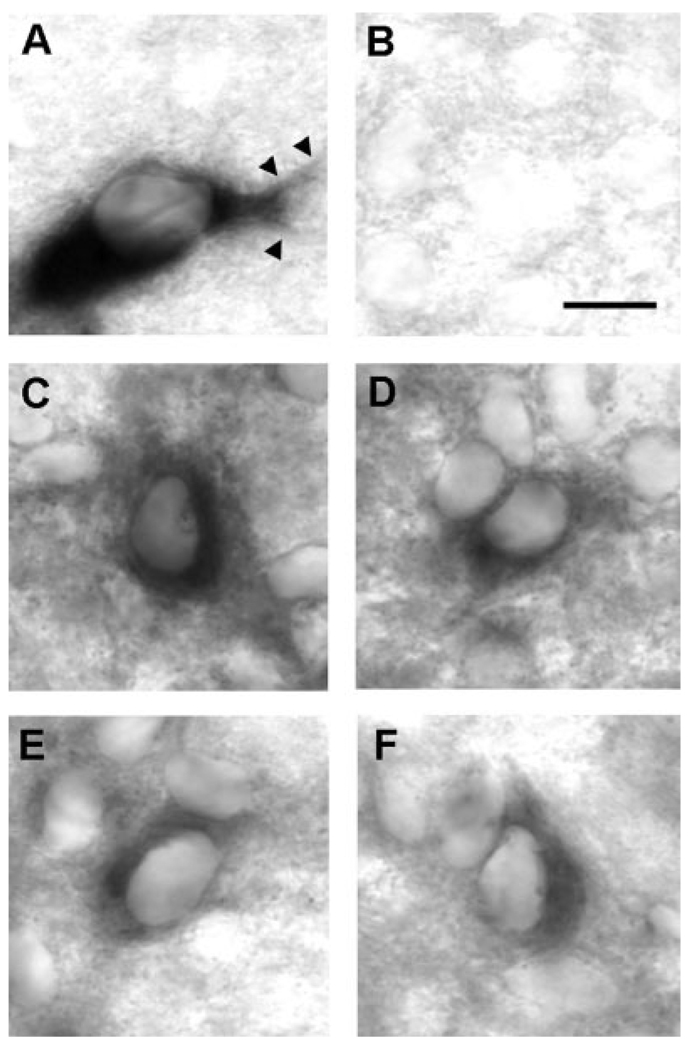
To determine the cellular basis of GDNF mRNA expression in striatum, we performed nonradioactive in situ hybridization (NRISH) on fixed, cryoprotected rat brains. Using this technique, deposition of chromogen throughout the cytoplasm of the cell soma and proximal dendrites makes possible a clear definition of cellular morphology, as demonstrated for an SN dopaminergic neuron using a riboprobe for the dopamine transporter (Fig. 2A). NRISH analysis of GDNF mRNA in striatum revealed labeled neuronal profiles at the cellular level (Fig. 2C–F). These profiles were distributed throughout the striatum in both the anterior-posterior and medial-lateral dimensions, although we noted that the most intensely labeled profiles tended to be more clustered laterally. An analysis of 163 labeled profiles observed among four striatal sections revealed that all of them had cellular morphologic features typical of neurons; abundant cytoplasm, a polygonal shape, and tapered proximal dendrites. There were no profiles that resembled astrocytes or microglia. The neurons observed in the striatum typically have a soma diameter of 10–15 µm, characteristic of medium-sized neurons, the most abundant type of the rat striatum (Carpenter, 1976).
Fig. 2.
Nonradioactive in situ hybridization staining of GDNF mRNA-positive profiles in striatum during development. A: A representative example of the cellular morphology defined by NRISH chromogen deposition is shown for a dopamine neuron of the SNpc by probing for the dopamine transporter (DAT). The cell soma is distinctly defined, and both primary and secondary dendrites (arrowheads) are demonstrated. B: A sense-GDNF probe reveals only faint background staining. C–F: Four examples of GDNF mRNA-positive neuronal profiles within the striatum at PND5. Scale bar = 10 µm.
GDNF protein expression in striatum
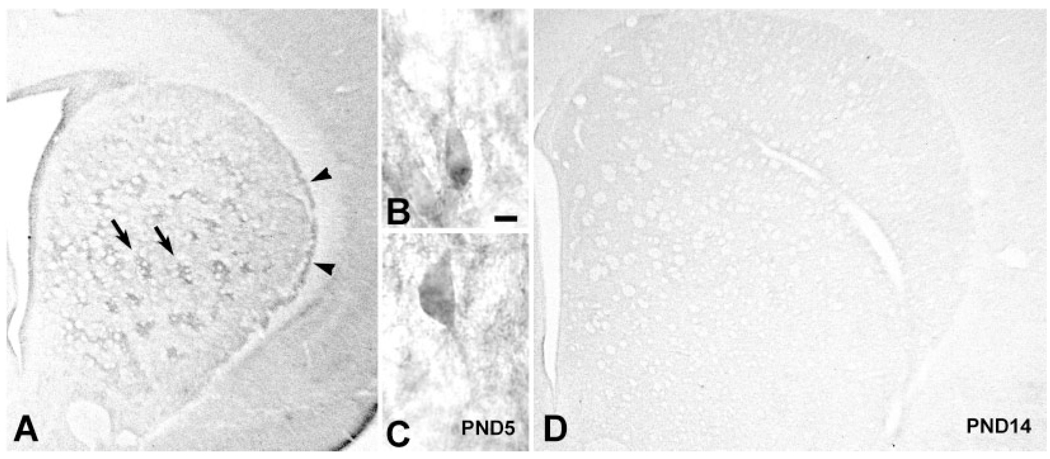
During the first postnatal week, striatal GDNF protein expression at a regional level was identified by immunoperoxidase staining in distinct patches (striosomes) and in a lateral crescent subjacent to the external capsule, as previously described (Lopez-Martin et al., 1999) (Fig. 3A). By the second postnatal week, staining had become faint and diffuse, and the distinct patch and lateral crescent staining was no longer observable (Fig. 3D). Thus, the period of most distinct immunostaining corresponds to the period of highest GDNF mRNA levels in the striatum (Cho et al., 2003). Therefore, further anatomical studies, as outlined below, were conducted in animals aged PND5–7.
Fig. 3.
Striatal GDNF immunoperoxidase staining during development. A: GDNF immunostaining at PND5 reveals distinctly stained patches (arrows) and a lateral crescent band of staining subjacent to the external capsule (arrowheads). B,C: Representative GDNF-positive neuronal profiles in striatum at PND5. The surrounding chromogen deposition is due to positive neuropil staining, out of the plane of focus. D: GDNF immunostaining at PND14 is diffuse and faint. Scale bar = 10 µm.
At a cellular level, rare GDNF-positive neuronal profiles were observed (Fig. 3B,C), confirming the NRISH finding of GDNF expression by intrinsic striatal neurons. However, these profiles were few in number and they did not cluster, so they did not account for the patches of staining observed at the regional level. As for the NRISH technique, we did not observe astrocyte or microglial morphology among GDNF immunopositive profiles.
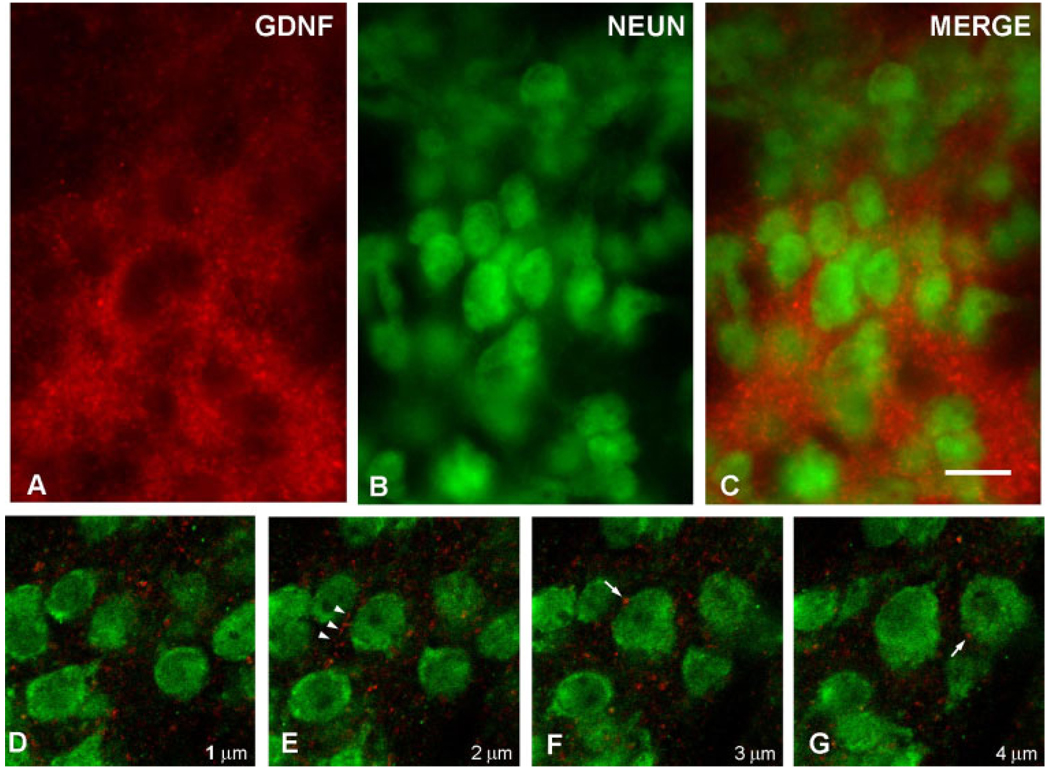
To further explore the cellular basis of the GDNF patch staining within the striatum during development, we performed immunofluorescence double-labeling for GDNF and the specific neuronal marker NeuN. Examination by epifluorescence revealed a dense matrix of fibrillar and punctate GDNF-positive structures enveloping NeuN-positive neuronal profiles (Fig. 4A–C). Examination of serial optical planes by confocal microscopy revealed numerous fibrillar and punctate structures adjacent to and sometimes abutting NeuN-positive profiles (Fig. 4D–G). In no instance did confocal analysis reveal GDNF positivity within the cytoplasm of a NeuN-positive profile within a striosome. We therefore conclude that the GDNF-positive striosomal patches observed during development consists predominantly of a fibrillar network.
Fig. 4.
Immunofluorescence labeling of GDNF and NeuN. A: A GDNF-positive striatal patch is identified by red AlexaFluor594 staining at PND6. Punctate and fibrillar staining is observed. B: Striatal neurons are identified by NeuN staining using a fluorescein label. C: A merged image reveals the NeuN-positive neurons enmeshed within the GDNF-positive fibrillar network. The extensive, overlapping GDNF fiber staining in this section observed by epifluorescence obscures any possible underlying GDNF staining within cytoplasm. Therefore, confocal analysis of serial optical sections was performed, as shown in consecutive sections D–G. These sections reveal fibrillar GDNF staining (arrowheads in E) and diffuse punctuate staining, some of it juxtaposed to neuronal cell bodies (arrows in F, G). No GDNF staining was observed in the cytoplasm of any NeuN-positive neurons in striosomes. The micron measurements in the lower right corner of panels D–G represent the Z-axis distances above the plane shown in D, arbitrarily defined as the initial plane at 1 µm. Scale bar = 10 µm in C (applies to A–G).
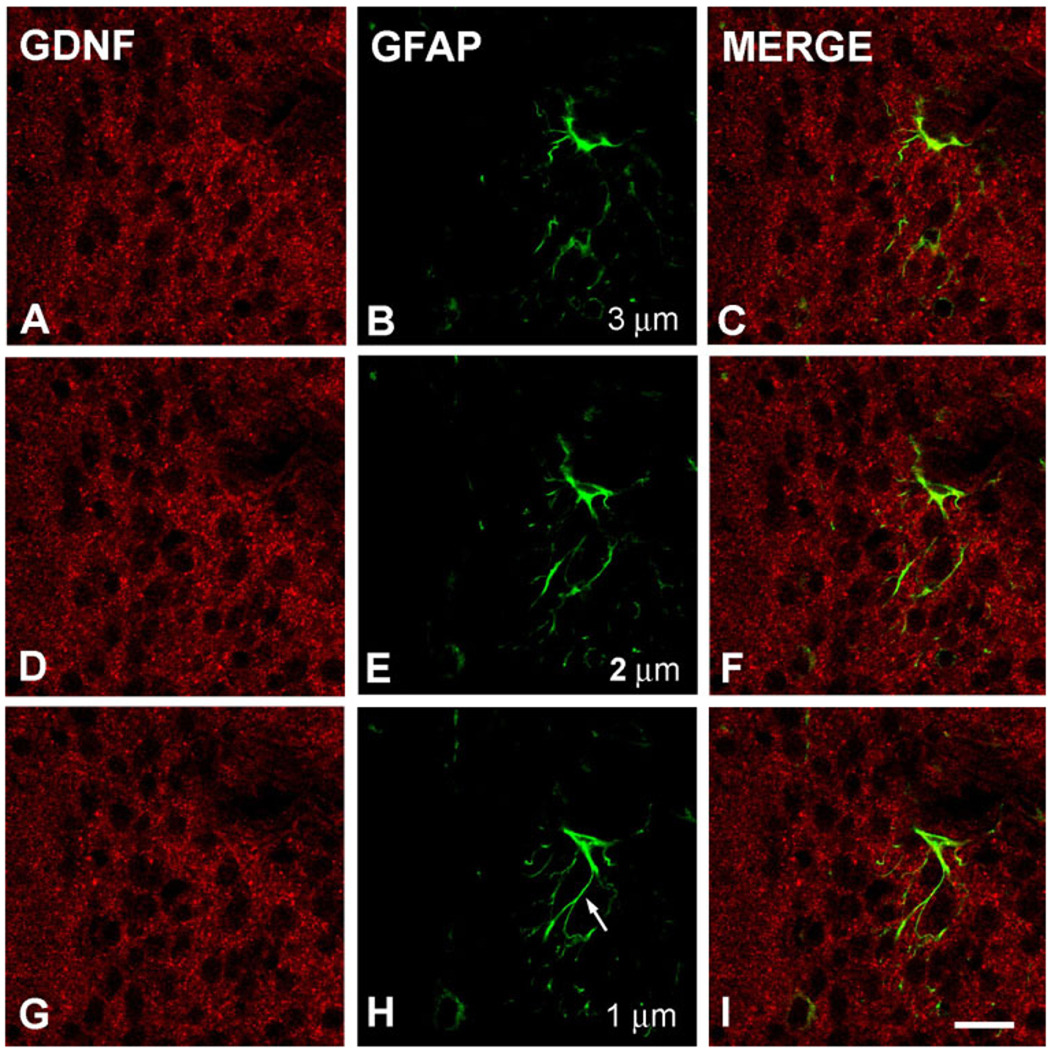
While the striosomal patch distribution pattern is an important and prevalent feature of the neuronal organization of the striatum (Graybiel, 1990), it has not been recognized as such for glial anatomical distribution. Nevertheless, we sought to evaluate any possible contribution by astrocytes to the GDNF-positive fibrillar matrix observed in striosomes, by performing double-labeling for GDNF and the astrocyte marker GFAP in n = 4 animals. At low power, there was no spatial relationship between GDNF patches and GFAP-positive astrocytes. Likewise, at high power, with analysis of serial optical planes by confocal microscopy, there was no apparent coregistration of GFAP-positive fiber staining with GDNF-positive fibers (Fig. 5).
Fig. 5.
Immunofluorescence labeling of GDNF and GFAP. Shown are three consecutive confocal planes, each labeled for GDNF (A,D,G) and GFAP (B,E,H) and merged (C,F,I) in a PND6 rat. The micron measurements in panels B, E, and H represent the Z-axis distances above the plane shown in H, arbitrarily defined as the initial plane at 1 µm. There is no cellular colocalization of the GDNF and GFAP staining. This is especially clear visually for the astrocyte fiber shown in panel H (arrow). Scale bar = 10 µm in I (applies to A–I).
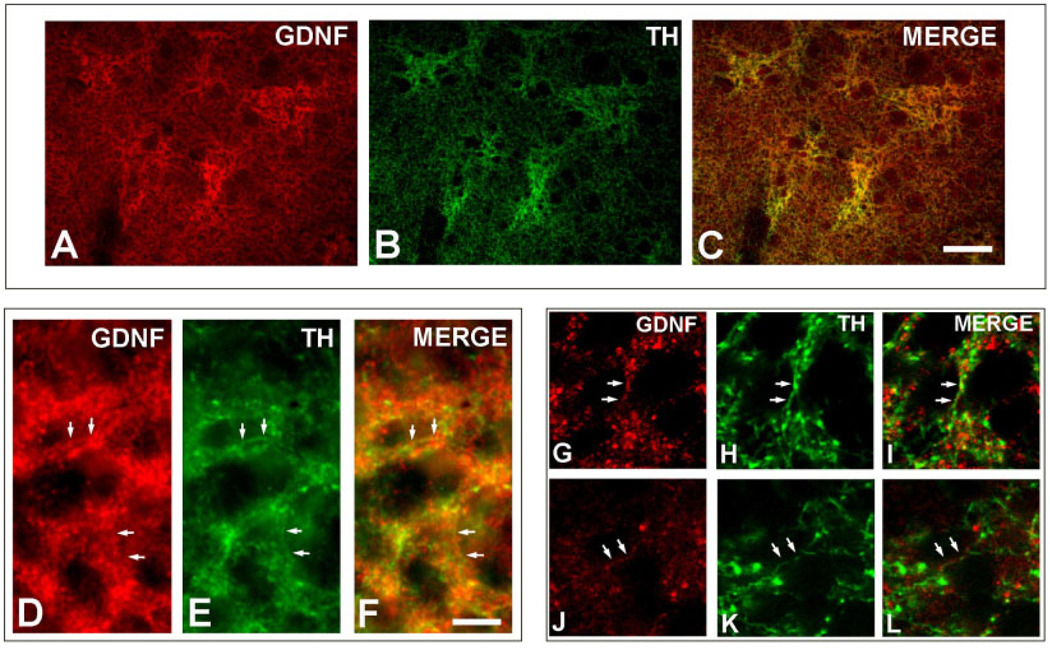
We considered the possibility that the GDNF-positive neuropil within striosomal patches may be derived from projecting afferent systems as well as fibers emanating from intrinsic striatal neurons. For the dopaminergic projection in particular, Lopez-Martin et al. (1999) had previously demonstrated a coregistration of TH-positive fibers with GDNF-positive striosomes. Therefore, to explore the relationship between GDNF striosomal fiber staining and TH-positive dopaminergic afferent fibers, we performed immunofluorescence double-labeling. At a regional level, this analysis revealed an exact coregistration of GDNF-positive striosomes with TH-positive striosomes (Fig. 6A–C). At a cellular level, epifluorescence microscopy revealed numerous instances of colabeling of fibers (Fig. 6D–F) and this was confirmed by confocal analysis (Fig. 6G–L). Although many instances of colocalization of GDNF and TH in striosomal fibers were readily identified, they were the minority of GDNF-positive fibers. While some of these GDNF-positive fibers may be negative for TH for technical reasons (e.g., related to tissue penetration of reagents), it is also quite likely that many of these GDNF-positive neurites arise from nondopaminergic neurons, either intrinsic striatal neurons or other afferent projections.
Fig. 6.
Immunofluorescence labeling of GDNF and TH. A lower-power epifluorescence micrograph demonstrates a correspondence between striosomes identified by GDNF staining (A) and those identified by TH staining (B), as shown in the merged image (C). At higher power, epifluorescence demonstrates colocalization of GDNF (D) and TH (E) in striosomal fibers (merged in F) (examples of colocalization are indicated by pairs of white arrows). Colocalization is confirmed by confocal analysis. Two representative examples are shown in panel sets G–I and J–L. Scale bars = 100 µm in C (applies to A–C); 10 µm in F (applies to D–L).
GDNF protein expression in striatal efferent targets
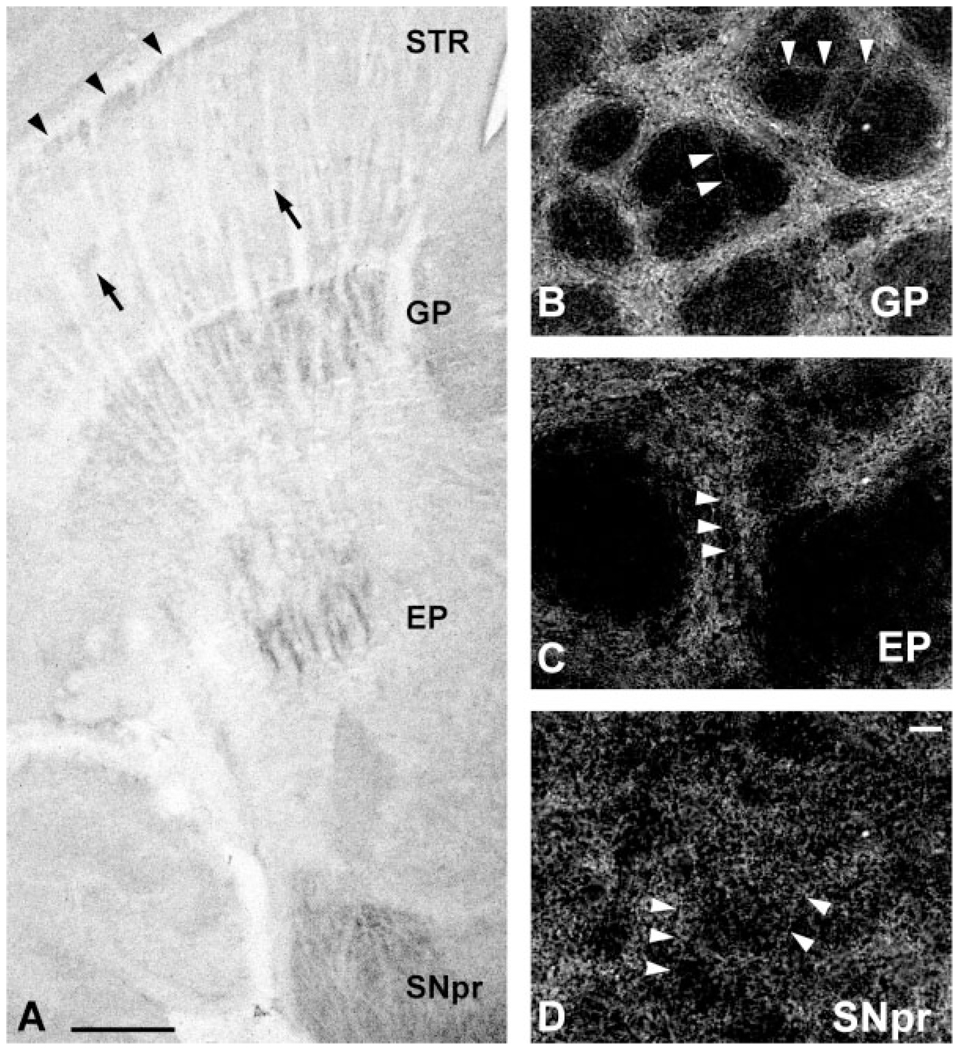
If GDNF protein is synthesized by intrinsic striatal neurons, then it would be predicted that it may be transported not only to local striatal neuropil, but also to the major efferent target nuclei of striatal neurons. The major striatal efferent targets in rodent are the globus pallidus (GP), the entopeduncular nucleus (EP), and the substantia nigra pars reticulata (SNpr) (Carpenter, 1976). We therefore examined immunostaining in these structures in PND5–7 animals. In order to compare levels of staining in these structures to that of the striatum, we performed staining on horizontal sections which included all four regions. At low power, striosomal and lateral crescent staining could be observed within the striatum (Fig. 7A). On these same sections, staining of a greater intensity than that observed within the striatum was demonstrated in the GP, EP, and the SNpr (Fig. 7A). The intensity of staining within these structures was comparable. At a cellular level the GP, EP, and SNpr all demonstrated numerous GDNF-positive fibers (Fig. 7B–D). In none of these structures were GDNF-positive profiles identified, and in the SNpr in particular, no stained cell bodies were observed. We therefore concluded that, as predicted from the presence of GDNF mRNA and protein in striatal intrinsic neurons, GDNF can be identified in fibers within the main striatal efferent targets. We also investigated patterns of GDNF regional staining in striatal efferent target structures at PND14. In these sections, immunostaining was discerned in the GP, EP, and SNpr, as ob- served in animals at PND5–7. However, as observed in the striatum at PND14 (Fig. 3D), it was faint and barely detectable above background (not shown).
Fig. 7.
Immunoperoxidase staining of GDNF in striatal efferent targets. A: A horizontal section immunoperoxidase stained for GDNF in PND6 rat. Faint striosomes (arrows) and the lateral crescent (arrowheads) can be observed in the striatum (STR). Distinct peroxidase staining is observed in globus pallidus (GP), entopeduncular nucleus (EP), and SNpr. At a cellular level, here shown in coronal sections by dark-field examination, numerous GDNF-positive fibers are observed in GP (B), EP (C), and SNpr (D). In each panel, examples of fibers are labeled with arrowheads. In none of these structures were cell bodies identified. Likewise, in SNpc, adjacent to the SNpr, cell bodies were not identified. Scale bars = 500 in A; 10 µm in D (applies to B–D).
DISCUSSION
The main goal of these investigations was to establish the cellular phenotype(s) responsible for the local production of GDNF in the striatum during early postnatal development, when it serves as a striatal target-derived neurotrophic factor for the regulation of the natural cell death event in SN dopamine neurons (Oo et al., 2003; Kholodilov et al., 2004). Since neuronal cellular proteins may be transported to distant axonal compartments, or, in the case of neurotrophic factors, released extracellularly, we anticipated that examination of GDNF mRNA would provide the best guide to cells responsible for local striatal production. In order to examine mRNA in a way that would permit clear examination of cellular morphology, we used a nonradioisotopic method in combination with tissue preservation by intracardiac perfusion fixation, and cryoprotection, as previously described for studies of GDNF mRNA expression in the adult brain (Pochon et al., 1997). This technique is compatible with clear examination of cellular morphology, because of the extensive and exclusive deposition of chromogen product within the cytoplasm of the cell soma. Our analysis demonstrated that GDNF mRNA is expressed within neurons of the striatum during postnatal development. These neurons were 10–15 µm in size, and thus belong to the medium-size class of striatal neurons, the most abundant class in the striatum (Carpenter, 1976). Thus, our analysis of the cellular phenotype responsible for striatal GDNF mRNA expression during development is similar to that of Pochon et al. (1997), performed on adult animals, who noted GDNF mRNA expression exclusively in neurons. While we can safely conclude that GDNF mRNA is expressed within medium-sized striatal neurons, we recognize that it may also be expressed in other neuronal and nonneuronal cellular phenotypes at levels below the limit of sensitivity of this technique.
To further explore the cellular origin of GDNF in the postnatal striatum, we performed immunohistochemistry. While few cellular profiles were identified, they were neuronal, in keeping with the results of the mRNA studies. A previous study of GDNF protein cellular expression in adult rats failed to reveal staining in normal animals, but demonstrated positive staining exclusively in neurons following ischemia (Abe and Hayashi, 1997). Having demonstrated abundant GDNF mRNA-positive neuronal profiles in striatum, we expected to observe a similar number by immunostaining for protein. While immunostaining demonstrated abundant neuropil, there were very few positive cell bodies, and their staining was faint. The explanation for this discrepancy is unknown, but one possible explanation is that after GDNF protein is synthesized within the cell body, it is rapidly transported into axons, or released extracellularly, or both.
Although we were unable to demonstrate the expression of GDNF mRNA or protein in glial cell bodies, we nevertheless considered the possibility that some of the GDNF-positive fiber staining demonstrated by the immunoperoxidase technique may be glial in origin. To assess this possibility, we examined tissue sections double-labeled for GDNF and GFAP by both epifluorescence and confocal microscopy. These studies also failed to reveal any relationship between GDNF protein expression and astrocytes. We therefore conclude that although GDNF was originally derived from a glial cell line, and although it can be expressed by glia in the context of brain injury (Batchelor et al., 1999), neither its mRNA nor its protein can be detected in glia in the striatum during normal postnatal development by methods which readily detect expression in medium-sized neurons.
In keeping with our conclusion that medium-sized striatal neurons produce GDNF during postnatal development, we observed GDNF by immunoperoxidase stain in fibers and axon terminals in the principal striatal efferent target nuclei: the GP, EP, and SNpr. The functional significance of this expression is not known, but it suggests the possibility that GDNF of striatal origin may not only provide trophic support in retrograde fashion to afferent projection systems, such as the nigrostriatal system, but also in anterograde fashion to striatal efferent targets. There are numerous precedents for such anterograde delivery of trophic support during development (Linden, 1994). The anterograde transport of GDNF along neuronal projections from the striatum to efferent targets, such as the SNpr, has also been demonstrated by experiments utilizing viral vectors to transduce intrinsic striatal neurons (Georgievska et al., 2004).
The principal morphologic feature of GDNF immunostaining in the postnatal striatum at the regional level is its prominent appearance in striosomal patches, as originally described by Lopez-Martin et al. (1999) and confirmed herein. We have shown in this analysis that the cellular basis of this striosomal pattern of staining is exclusively GDNF-positive neuropil; there is no significant contribution of cell body staining. Given the local production of GDNF mRNA and protein by medium-sized intrinsic striatal neurons, and given that GDNF-positive fibers were observed in known striatal neuron projection systems, it is likely that this neuropil derives, at least in part, from local intrinsic striatal neurons. However, other anatomical systems also make a contribution, as we have shown that GDNF-positivity within striosomes could be demonstrated within TH-positive fibers arising from the nigro-striatal dopaminergic system.
We interpret our anatomical demonstration of GDNF immunoreactivity within striatal TH-positive fibers to represent GDNF undergoing retrograde transport following local production in the striatum and uptake by dopaminergic terminals. While we have not performed a functional analysis to formally exclude anterograde transport following local production in SNpc, that explanation is not favored by the weight of the evidence. We have shown that GDNF mRNA is more abundant in striatum than SN, and while GDNF-positive cell body immunostaining can be demonstrated in striatum during development, it cannot in SNpc. On the other hand, many investigations have demonstrated that the principal GDNF receptor, GFRα1, is far more abundant in SNpc than striatum (Widenfalk et al., 1997; Yu et al., 1998; Nosrat et al., 1997; Golden et al., 1998; Glazner et al., 1998; Horger et al., 1998). Furthermore, the capability of the nigro-striatal dopaminergic system to specifically transport GDNF retrograde has been demonstrated (Tomac et al., 1995b). Thus, the available evidence would suggest that GDNF in striatal TH-positive terminals is more likely to be undergoing retrograde transport. This interpretation notwithstanding, we have demonstrated the presence of GDNF mRNA in SN both by Northern analysis and NRISH (data not shown). It is therefore possible that GDNF is also locally produced in SN and serves an autocrine or paracrine role.
In summary, we have demonstrated that medium-sized striatal neurons express GDNF mRNA and protein during the first postnatal week, and we conclude that they are the source of the striatal GDNF which regulates the natural cell death event in SNpc dopamine neurons during this developmental period (Oo et al., 2003; Kholodilov et al., 2004). In accordance with such a neurotrophic role for GDNF, we have directly demonstrated its presence within striatal TH-positive dopaminergic fibers, and we interpret the available data on GDNF and GFRa1 expression to suggest that GDNF is undergoing retrograde transport in these fibers. In addition, we demonstrate that GDNF protein is localized not only to local striatal neuropil, but also fibers within striatal efferent targets. Thus, GDNF may play a role in regulating development not only in the dopaminergic nigro-striatal projection, but also in other structures with close anatomical relationships to the striatum.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant number: NS26836; NS38370; Grant sponsor: U.S. Department of Defense; Grant number DAMD17-03-1-0492; Grant sponsor: Parkinson’s Disease Foundation.
LITERATURE CITED
- Abe K, Hayashi T. Expression of the glial cell line-derived neurotrophic factor gene in rat brain after transient MCA occlusion. Brain Res. 1997;776:230–234. doi: 10.1016/s0006-8993(97)01041-x. [DOI] [PubMed] [Google Scholar]
- Batchelor PE, Liberatore GT, Wong JY, Porritt MJ, Frerichs F, Donnan GA, Howells DW. Activated macrophages and microglia induce dopaminergic sprouting in the injured striatum and express brain- derived neurotrophic factor and glial cell line-derived neurotrophic factor. J Neurosci. 1999;19:1708–1716. doi: 10.1523/JNEUROSCI.19-05-01708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya B, Mandal C, Basu S, Sarkar PK. Regulation of alpha-and beta-tubulin mRNAs in rat brain during synaptogenesis. Brain Res. 1987;388:159–162. doi: 10.1016/s0006-8993(87)80009-4. [DOI] [PubMed] [Google Scholar]
- Blum M, Weickert CS. GDNF mRNA expression in normal postnatal development, aging, and in weaver mutant mice. Neurobiol Aging. 1995;16:925–929. doi: 10.1016/0197-4580(95)02011-x. [DOI] [PubMed] [Google Scholar]
- Bowenkamp KE, Hoffman AF, Gerhardt GA, Henry MA, Biddle PT, Hoffer BJ, Granholm A-CE. Glial cell line-derived neurotrophic factor supports survival of injured midbrain dopaminergic neurons. J Comp Neurol. 1995;355:479–489. doi: 10.1002/cne.903550402. [DOI] [PubMed] [Google Scholar]
- Burke RE, Franklin SO, Inturrisi CE. Acute and persistent suppression of preproenkephalin mRNA expression in the striatum following developmental hypoxic-ischemic injury. J Neurochem. 1994;62:1878–1886. doi: 10.1046/j.1471-4159.1994.62051878.x. [DOI] [PubMed] [Google Scholar]
- Burke RE, Antonelli M, Sulzer D. Glial cell line-derived neurotrophic growth factor inhibits apoptotic death of postnatal substantia nigra dopamine neurons in primary culture. J Neurochem. 1998;71:517–525. doi: 10.1046/j.1471-4159.1998.71020517.x. [DOI] [PubMed] [Google Scholar]
- Carpenter MB. Anatomical organization of the corpus striatum and related nuclei. In: Yahr MD, editor. The basal ganglia. New York: Raven Press; 1976. pp. 1–36. [PubMed] [Google Scholar]
- Cho J, Kholodilov NG, Burke RE. The developmental time course of glial cell line-derived neurotrophic factor (GDNF) and GDNF receptor alpha-1 mRNA expression in the striatum and substantia nigra. Ann N Y Acad Sci. 2003;991:284–287. [Google Scholar]
- Choi-Lundberg DL, Bohn MC. Ontogeny and distribution of glial cell line-derived neurotrophic factor (GDNF) mRNA in rat. Dev Brain Res. 1995;85:80–88. doi: 10.1016/0165-3806(94)00197-8. [DOI] [PubMed] [Google Scholar]
- Choi-Lundberg DL, Lin Q, Chang YN, Chiang YL, Hay CM, Mohajeri H, Davidson BL, Bohn MC. Dopaminergic neurons protected from degeneration by GDNF gene therapy. Science. 1997;275:838–841. doi: 10.1126/science.275.5301.838. [DOI] [PubMed] [Google Scholar]
- El-Khodor BF, Kholodilov NG, Yarygina O, Burke RE. The expression of mRNAs for the proteasome complex is developmentally regulated in the rat mesencephalon. Brain Res Dev Brain Res. 2001;129:47–56. doi: 10.1016/s0165-3806(01)00181-x. [DOI] [PubMed] [Google Scholar]
- Gash DM, Zhang ZM, Ovadia A, Cass WA, Yi A, Simmerman L, Russell D, Martin D, Lapchak PA, Collins F, Hoffer BJ, Gerhardt GA. Functional recovery in parkinsonian monkeys treated with GDNF. Nature. 1996;380:252–255. doi: 10.1038/380252a0. [DOI] [PubMed] [Google Scholar]
- Georgievska B, Kirik D, Bjorklund A. Aberrant sprouting and down- regulation of tyrosine hydroxylase in lesioned nigrostriatal dopamine neurons induced by long-lasting overexpression of glial cell line derived neurotrophic factor in the striatum by lentiviral gene transfer. Exp Neurol. 2002;177:461–474. doi: 10.1006/exnr.2002.8006. [DOI] [PubMed] [Google Scholar]
- Georgievska B, Kirik D, Bjorklund A. Overexpression of glial cell line-derived neurotrophic factor using a lentiviral vector induces time- and dose-dependent downregulation of tyrosine hydroxylase in the intact nigrostriatal dopamine system. J Neurosci. 2004;24:6437–6445. doi: 10.1523/JNEUROSCI.1122-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Patel NK, Hotton GR, O’Sullivan K, McCarter R, Bunnage M, Brooks DJ, Svendsen CN, Heywood P. Direct brain infusion of glial cell line-derived neurotrophic factor in Parkinson disease. Nat Med. 2003;9:589–595. doi: 10.1038/nm850. [DOI] [PubMed] [Google Scholar]
- Glazner GW, Mu X, Springer JE. Localization of glial cell line-derived neurotrophic factor receptor alpha and c-ret mRNA in rat central nervous system. J Comp Neurol. 1998;391:42–49. doi: 10.1002/(sici)1096-9861(19980202)391:1<42::aid-cne4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Golden JP, Baloh RH, Kotzbauer PT, Lampe PA, Osborne PA, Milbrandt J, Johnson EMJ. Expression of neurturin, GDNF, and their receptors in the adult mouse CNS. J Comp Neurol. 1998;398:139–150. doi: 10.1002/(sici)1096-9861(19980817)398:1<139::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Golden JP, DeMaro JA, Osborne PA, Milbrandt J, Johnson EMJ. Expression of neurturin, GDNF, and GDNF family-receptor mRNA in the developing and mature mouse. Exp Neurol. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- Graybiel AM. Neurotransmitters and neuromodulators in the basal ganglia. Trends Neurosci. 1990;13:244–254. doi: 10.1016/0166-2236(90)90104-i. [DOI] [PubMed] [Google Scholar]
- Horger BA, Nishimura MC, Armanini MP, Wang LC, Poulsen KT, Rosen-blad C, Kirik D, Moffat B, Simmons L, Johnson EJ, Milbrandt J, Rosenthal A, Bjorklund A, Vandlen RA, Hynes MA, Phillips HS. Neurturin exerts potent actions on survival and function of midbrain dopaminergic neurons. J Neurosci. 1998;18:4929–4937. doi: 10.1523/JNEUROSCI.18-13-04929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Lewis V, Vila M, Djaldetti R, Guegan C, Liberatore G, Liu J, O’Malley KL, Burke RE, Przedborski S. Developmental cell death in dopaminergic neurons of the substantia nigra of mice. J Comp Neurol. 2000;424:476–488. doi: 10.1002/1096-9861(20000828)424:3<476::aid-cne6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Janec E, Burke RE. Naturally occurring cell death during postnatal development of the substantia nigra of the rat. Mol Cell Neurosci. 1993;4:30–35. doi: 10.1006/mcne.1993.1004. [DOI] [PubMed] [Google Scholar]
- Kholodilov N, Yarygina O, Oo TF, Zhang H, Sulzer D, Dauer WT, Burke RE. Regulation of the development of mesencephalic dopaminergic systems by the selective expression of glial cell line-derived neurotrophic factor in their targets. J Neurosci. 2004;24:3136–3146. doi: 10.1523/JNEUROSCI.4506-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Bjorklund A, Mandel RJ. Long-term rAAV- mediated gene transfer of GDNF in the rat Parkinson’s model: intra- striatal but not intranigral transduction promotes functional regeneration in the lesioned nigrostriatal system. J Neurosci. 2000;20:4686–4700. doi: 10.1523/JNEUROSCI.20-12-04686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordower JH, Emborg ME, Bloch J, Ma SY, Chu Y, Leventhal L, McBride J, Chen EY, Palfi S, Roitberg BZ, Brown WD, Holden JE, Pyzalski R, Taylor MD, Carvey P, Ling Z, Trono D, Hantraye P, Deglon N, Aebi- scher P. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson’s disease. Science. 2000;290:767–773. doi: 10.1126/science.290.5492.767. [DOI] [PubMed] [Google Scholar]
- Lazarini F, Deslys JP, Dormont D. Regulation of the glial fibrillary acidic protein, beta actin and prion protein mRNAs during brain development in mouse. Brain Res Mol Brain Res. 1991;10:343–346. doi: 10.1016/0169-328x(91)90093-d. [DOI] [PubMed] [Google Scholar]
- Lin L-FH, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- Linden R. The survival of developing neurons: a review of afferent control. Neuroscience. 1994;58:671–682. doi: 10.1016/0306-4522(94)90447-2. [DOI] [PubMed] [Google Scholar]
- Lopez-Martin E, Caruncho HJ, Rodriguez-Pallares J, Guerra MJ, Labandeira-Garcia JL. Striatal dopaminergic afferents concentrate in GDNF-positive patches during development and in developing intrastriatal striatal grafts. J Comp Neurol. 1999;406:199–206. doi: 10.1002/(sici)1096-9861(19990405)406:2<199::aid-cne5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Vallee A, Beaulieu C, Herman IM, Leclerc N. beta- Actin is confined to structures having high capacity of remodelling in developing and adult rat cerebellum. Eur J Neurosci. 1998;10:3785–3798. doi: 10.1046/j.1460-9568.1998.00391.x. [DOI] [PubMed] [Google Scholar]
- Nosrat CA, Tomac A, Hoffer BJ, Olson L. Cellular and developmental patterns of expression of Ret and glial cell line-derived neurotrophic factor receptor alpha mRNAs. Exp Brain Res. 1997;115:410–422. doi: 10.1007/pl00005711. [DOI] [PubMed] [Google Scholar]
- Oo TF, Burke RE. The time course of developmental cell death in phenotypically defined dopaminergic neurons of the substantia nigra. Dev Brain Res. 1997;98:191–196. doi: 10.1016/s0165-3806(96)00173-3. [DOI] [PubMed] [Google Scholar]
- Oo TF, Kholodilov N, Burke RE. Regulation of natural cell death in dopaminergic neurons of the substantia nigra by striatal GDNF in vivo. J Neurosci. 2003;23:5141–5148. doi: 10.1523/JNEUROSCI.23-12-05141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina SM, Quartu M, Ambu R, Follesa P, Del Fiacco M. Immunohis- tochemical localization of GDNF in the human hippocampal formation from prenatal life to adulthood. Brain Res. 2002;928:138–146. doi: 10.1016/s0006-8993(01)03377-7. [DOI] [PubMed] [Google Scholar]
- Pochon NA, Menoud A, Tseng JL, Zurn AD, Aebischer P. Neuronal GDNF expression in the adult rat nervous system identified by in situ hybridization. Eur J Neurosci. 1997;9:463–471. doi: 10.1111/j.1460-9568.1997.tb01623.x. [DOI] [PubMed] [Google Scholar]
- Sauer H, Rosenblad C, Bjorklund A. Glial cell line-derived neurotrophic factor but not transforming growth factor β3 prevents delayed degeneration of nigral dopaminergic neurons following striatal 6-hydroxydopamine lesion. Proc Natl Acad Sci U S A. 1995;92:8935–8939. doi: 10.1073/pnas.92.19.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar DG, Sieber BA, Dreyfus CF, Black IB. Regional and cell specific expression of GDNF in rat brain. Exp Neurol. 1993;124:368–371. doi: 10.1006/exnr.1993.1207. [DOI] [PubMed] [Google Scholar]
- Stromberg I, Bjorklund L, Johansson M, Tomac A, Collins F, Olson L, Hoffer B, Humpel C. Glial cell line derived neurotrophic factor is expressed in the developing but not adult striatum and stimulates developing dopamine neurons in vivo. Exp Neurol. 1993;124:401–412. doi: 10.1006/exnr.1993.1214. [DOI] [PubMed] [Google Scholar]
- Tomac A, Lindqvist E, Lin L-FH, Ogren SO, Young D, Hoffer BJ, Olson L. Protection and repair of the nigrostriatal dopaminergic system by GDNF in vivo. Nature. 1995a;373:335–339. doi: 10.1038/373335a0. [DOI] [PubMed] [Google Scholar]
- Tomac A, Widenfalk J, Lin LH, Kohno T, Ebendal T, Hoffer BJ, Olson L. Retrograde axonal transport of glial cell line-derived neurotrophic factor in the adult nigrostriatal system suggests a trophic role in the adult. Proc Natl Acad Sci U S A. 1995b;92:8274–8278. doi: 10.1073/pnas.92.18.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupp M, Belluardo N, Funakoshi H, Ibanez CF. Complementary and overlapping expression of glial cell line-derived neurotrophic factor (GDNF), c-ret proto-oncogene, and GDNF receptor-alpha indicates multiple mechanisms of trophic actions in the adult rat CNS. J Neurosci. 1997;17:3554–3567. doi: 10.1523/JNEUROSCI.17-10-03554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widenfalk J, Nosrat C, Tomac A, Westphal H, Hoffer B, Olson L. Neurturin and glial cell line-derived neurotrophic factor receptor-beta (GDNFR-beta), novel proteins related to GDNF and GDNFR-alpha with specific cellular patterns of expression suggesting roles in the developing and adult nervous system and in peripheral organs. J Neurosci. 1997;17:8506–8519. doi: 10.1523/JNEUROSCI.17-21-08506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Scully S, Yu Y, Fox GM, Jing S, Zhou R. Expression of GDNF family receptor components during development: implications in the mechanisms of interaction. J Neurosci. 1998;18:4684–4696. doi: 10.1523/JNEUROSCI.18-12-04684.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]