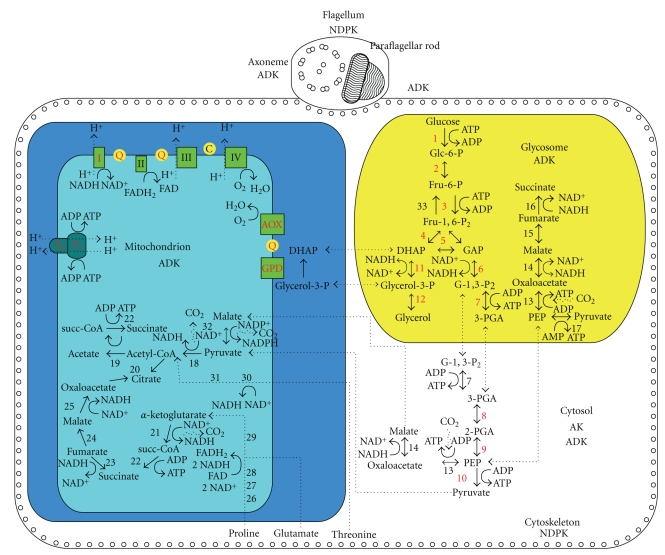
Figure 1.
Schematic representation of trypanosomatids' energy metabolism. Enzymes are indicated with numbers: 1, hexokinase; 2, glucose-6-phosphate isomerase; 3, phosphofructokinase; 4, aldolase; 5, triosephosphate isomerase; 6, glyceraldehyde-3-phosphate dehydrogenase; 7, phosphoglycerate kinase; 8, phosphoglycerate mutase; 9, enolase; 10, pyruvate kinase; 11, glycerol-3-phosphate dehydrogenase; 12, glycerol kinase; 13, phosphoenolpyruvate carboxykinase; 14, malate dehydrogenase; 15, fumarase; 16, fumarate reductase; 17, pyruvate phosphate dikinase; 18, pyruvate dehydrogenase complex; 19, acetate:succinate CoA transferase; 20, citrate synthase; 21, α-Ketoglutarate dehydrogenase; 22, succinyl-CoA synthetase; 23, succinate dehydrogenase; 24, fumarase; 25, malate dehydrogenase; 26, proline oxidase; 27, Δ′-pyrroline-5-carboxylate reductase; 28, glutamate semialdehyde dehydrogenase; 29, glutamate dehydrogenase; 30, threonine dehydrogenase; 31, acetyl-CoA : glycine C-acetyltransferase; 32, NADP-linked decarboxylating malic enzyme; 33, fructose-1,6-bisphosphatase. Enzymes present in bloodstream forms only are indicated in red, procyclic forms enzymes are in red and black. AOX: alternative oxidase; GPD: FAD-dependent glycerol-3-phosphate dehydrogenase; I: NADH-ubiquinone oxidoreductase (complex I); II: succinate dehydrogenase (complex II); III: cytochrome c reductase (complex III); IV: cytochrome c oxidase (complex IV); c: cytochrome c; Q: ubiquinone; F0/F1: F0/F1-ATP synthase; Glc-6-P: glucose-6-phosphate; Fru-6-P: fructose-6-phosphate; Fru-1,6-P2: fructose-1,6-bisphosphate; GAP: glyceraldehyde-3-phosphate; G-1,3-P2: 1,3-bisphosphoglycerate; 3-PGA: 3-phosphoglycerate; 2-PGA: 2-phosphoglycerate; PEP: phosphoenolpyruvate; DHAP: dihydroxyacetone phosphate; glycerol-3-P: glycerol-3-phosphate; succ-CoA: succinyl-coenzyme A; acetyl-CoA, acetyl-coenzyme A. For a detailed explanation see the text.