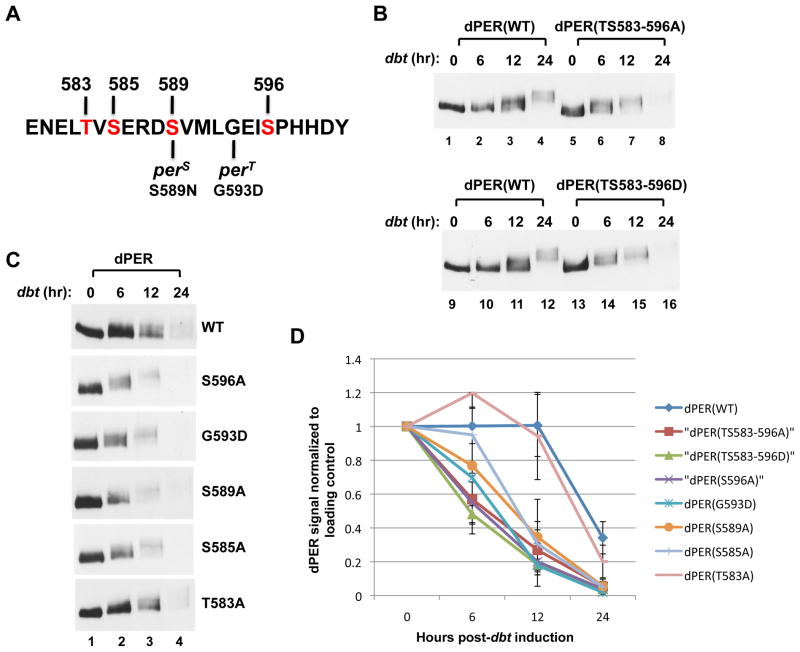
Figure 1. Differential effects of phospho-site mutations in the perS phospho-cluster on DBT-dependent phosphorylation kinetics and degradation rate of dPER in cultured S2 cells.
(A) Sequence of dPER from amino acid 579 to 601, encompassing the original “per-short” region (aa 585 to 601), highlighting the phosphorylation site mutations generated in this study (labeled in red) as well as locations of the perS and perT mutations. (B, C) S2 cells were co-transfected with wild type (WT) or mutant variants of pAc-3XFLAG-His-dper-6Xcmyc (simplified as dper) and pMT-dbt(WT)-V5-His (simplified as dbt, left of panels), and collected at the indicated times (hr) post-dbt induction. The dPER variants (top of panels) were detected by immunoblotting in the presence of α-c-myc antibodies. (D) dPER staining intensity values normalized to Hsp70 loading control. At least two independent experiments were used to generate the average for each time point. Error bars represent SEM. See Fig. S1 for supporting data.