Figure 1.
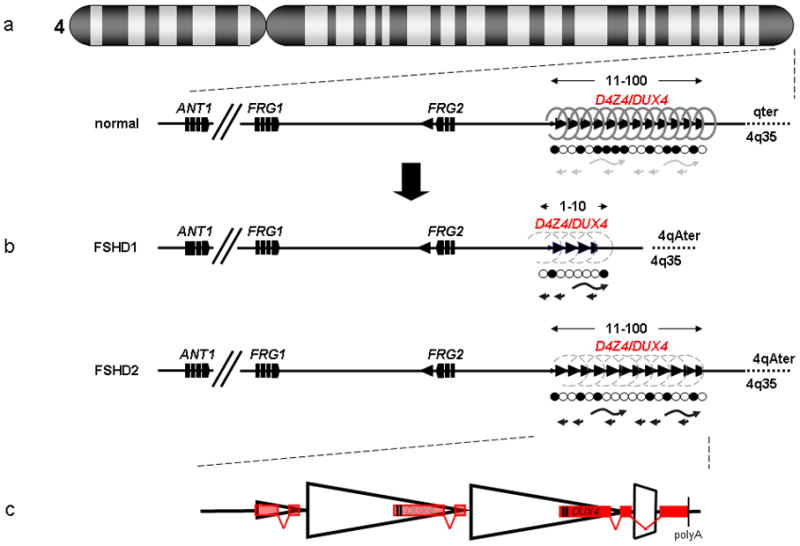
Schematic of the FSHD locus. (a) The D4Z4 repeat (triangles) is located in the subtelomere of chromosome 4q and can vary between 11–100 copies in the unaffected population. This repeat structure has a closed chromatin structure characterized by heterochromatic histone modifications (dense springs), high DNA methylation levels (closed circles) and complex bidirectional transcriptional activity (grey arrows). Candidate genes DUX4, FRG2, FRG1 and ANT1 are indicated. (b) In patients with FSHD, the chromatin structure of D4Z4 adopts a more open configuration (open springs and open circles) leading to inefficient transcriptional repression (black arrows) of the D4Z4 repeat. (c) The DUX4 gene is located within each D4Z4 unit. On permissive chromosomes, the last copy of the DUX4 genes splices to a third exon located in the region immediately flanking the repeat and stabilizing the transcript due to the presence of a polyadenylation (polyA) signal. Figure modified, with permission, from [52].
