Abstract
The retinoblastoma protein (Rb) regulates cell proliferation through its association with E2F transcription factors and other proteins. The Rb “pocket” domain primarily facilitates protein-protein interactions, and several structures of the pocket bound to E2F and tumorigenic viral proteins have been reported. We report here the first crystal structure of the pocket domain without bound ligand. We find that ligand association results in observable structural changes at the binding sites but no significant changes to the overall conformation of the domain. These data support models for regulation of Rb-E2F binding that do not require considerable structural changes in the pocket domain.
Keywords: retinoblastoma protein, tumor-suppressor protein, cell cycle, transcription regulation, protein-protein interactions, x-ray structure
Introduction
The retinoblastoma gene product (Rb) is a tumor suppressor protein thought to be inactivated in the majority of human cancers.1,2 Rb controls diverse processes related to cell growth including the cell cycle, senescence and differentiation, apoptosis, and chromatin maintenance.1–3,4,5 The most extensively characterized function of Rb is its regulation of gene expression; Rb directly inhibits E2F transcription factors and recruits various chromatin modifying proteins to E2F promoters. Nearly all of the protein binding sites identified in Rb are located in the structured pocket domain and the intrinsically disordered C-terminal domain (RbC).5 Two binding sites in the pocket domain, the E2F transactivation domain (E2FTD) site and the so-called ‘LxCxE’ site, have been characterized through crystallographic studies.6–10 The LxCxE site is a target site for oncogenic viral proteins that disrupt Rb-E2F complexes and contain the LxCxE sequence motif. Many cellular proteins are also thought to bind at the LxCxE site, although there is no available structure of such a complex.
Rb is regulated by multisite phosphorylation, which promotes dissociation from its binding partners. Rb phosphorylation primarily occurs in unstructured regions and induces intramolecular associations between the phosphorylated sequences and the structured domains.11–13 How these phosphorylation-driven conformational changes dissociate E2F is a question addressed by ongoing research, and two distinct models have been proposed. In one model, binding of the pocket to phosphorylated RbC causes a conformational change in the pocket domain that exposes S567 for subsequent phosphorylation.12 S567 phosphorylation is thought to disrupt the pocket interface, resulting in loss of E2FTD binding. In a second model, interactions between phosphorylated sequences and the pocket directly inhibit transcription factor binding by blocking the pocket binding sites. A key unresolved difference between these models is whether the pocket undergoes a significant structural change upon binding the phosphorylated motifs.
Rb acts as an adaptor protein that assembles diverse multiprotein complexes on chromatin. A critical question remains how binding of multiple protein ligands to Rb is coordinated to produce distinct biological outputs. In most cases it is unclear whether ligand binding at one site influences interactions at other sites and whether this relies on allosteric structural effects. In order to understand how potential conformational changes in Rb contribute to ligand binding and phosphorylation induced ligand-dissociation, it is important to examine the structures of both bound and unliganded Rb. Presently there are two crystal structures of the Rb pocket domain bound to E2FTD peptides,7,10 two structures with viral proteins complexed at the LxCxE site,6,8 and a structure of a viral protein bound at the E2FTD site.9 Here we report the first crystal structure of the human Rb pocket domain in the absence of any ligand and present for the first time a complete structural analysis of the changes that occur in Rb upon binding E2FTD and LxCxE containing proteins. Our results also have implications regarding proposed mechanisms for Rb-E2F dissociation.
Results and Discussion
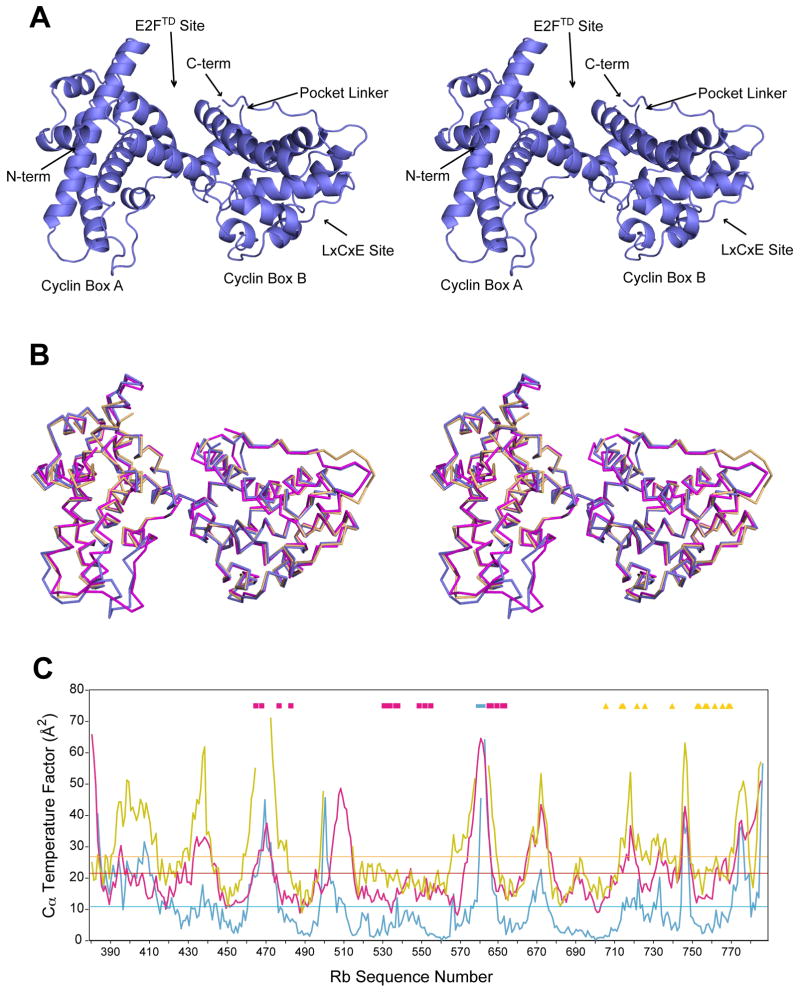
We found conditions in which crystals of the Rb pocket domain could be grown in the absence of any ligand and solved the structure to 2.5 Å resolution [Fig. 1(A)]. Statistics for data collection and refinement are presented in Table I. The asymmetric unit contains two copies of the molecule that are superimposable with an rmsd of 0.46 Å between 305 Cα atoms. As previously described, the pocket consists of two subdomains that are called the A and B cyclin boxes because each has a helical cyclin fold.8 The E2F transactivation domain binding site is at the interface between the A and B boxes, while the LxCxE binding site is in a cleft within the B box. There is a disordered linker of ~65 amino acids between the A and B subdomains, which must be deleted for crystallization.8 The pocket linker (residues 578–642) was removed in our engineered construct and replaced with a thrombin cleavage site, however crystallization was best achieved without using the protease. Small angle x-ray scattering (SAXS) data demonstrate that the conformation of the pocket domain with the thrombin site cleaved in solution is similar to the conformation of the refined crystallographic model, indicating that the artificially shortened linker does not perturb the structure (Supplemental Fig. 1).14
Figure 1.
Structural comparison of unliganded and liganded Rb pocket domains. (A) Ribbon diagram in stereoview for the unliganded Rb pocket domain shows the overall fold. (B) Superposition of the unliganded structure (blue) with the structure bound to E2FTD (pink, PDB ID: 1N4M) and bound to an LxCxE containing peptide from the human papilloma virus E7 protein (gold, PDB ID: 1GUX). (C) Crystallographic temperature factors for Cα atoms in the unliganded Rb pocket domain structure (blue), the E2FTD-bound structure (pink), and the E7-bound structure (gold). The mean Cα temperature factors for each refined structural model are indicated by the horizontal lines. Residues that compose the E2FTD (pink squares) and LxCxE (gold triangles) binding sites and pocket linker (blue bar) are marked.
Table I.
| Data Collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 60.28, 111.16, 138.93 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 60.3-2.5 |
| Rmerge (%) | 8.0 (38.7) |
| I/σI | 16.6 (3.9) |
| Completeness (%) | 99.9 (100.0) |
| Redundancy | 7.1 |
| Refinement | |
| Resolution (Å) | 2.5 |
| No. Reflections | 31,473 |
| Rwork/Rfree (%) | 21.3/26.2 |
| No. Atoms | |
| Total | 5727 |
| Protein | 5547 |
| Water | 180 |
| Average B-factor (Å2) | |
| Total | 16.2 |
| Protein | 16.2 |
| Water | 16.8 |
| R.m.s. Deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.532 |
| Ramachandran Analysis | |
| Most favored (%) | 89.7 |
| Additionally allowed | 10.0 |
| Generously allowed | 0.3 |
Values in parenthesis correspond to the highest resolution shell (2.64-2.50 Å)
The unliganded pocket structure has a similar overall conformation as previous crystal structures containing peptides at either of the two binding sites [Fig. 1(B)].7,8 We present here detailed comparisons with two representative ligand-bound structures, although similar observations were made with all available structures. Pairwise alignment with the E2FTD-bound pocket structure (PDB ID: 1N4M) results in an rmsd of 0.88 Å for 317 Cα atoms. Alignment with the structure of Rb bound to the LxCxE peptide from the human papilloma virus E7 protein (PDB ID: 1GUX) occurs with an rmsd of 0.53 Å between 292 Cα atoms. These similar conformations are likely not an artifact of crystal lattice constraints, as the packing interfaces in each crystal are distinct (Supplemental Fig. 2). Furthermore, SAXS data confirm that the overall conformation of the pocket domain in solution does not change with E2FTD binding (Supplemental Fig. 1).
The most significant differences in the structures are in the loops corresponding to residues 436–438, 500–513, and 772–777; however, these sequences have relatively high crystallographic temperature factors [Fig. 1(C)], suggesting they are dynamic. Although the positions of the more ordered secondary structure elements are nearly identical among the three structures, there are significant changes in helical position at the end of helix α4 and the beginning of helix α11, both of which tip towards the E2FTD peptide to facilitate interactions in that structure. Comparison of Cα temperature factors across the structures reveals few differences in values relative to each mean [Fig. 1(C)]. There are no regions that become more ordered upon ligand binding, which is consistent with the fact that that both binding surfaces are composed from residues primarily in ordered helices.
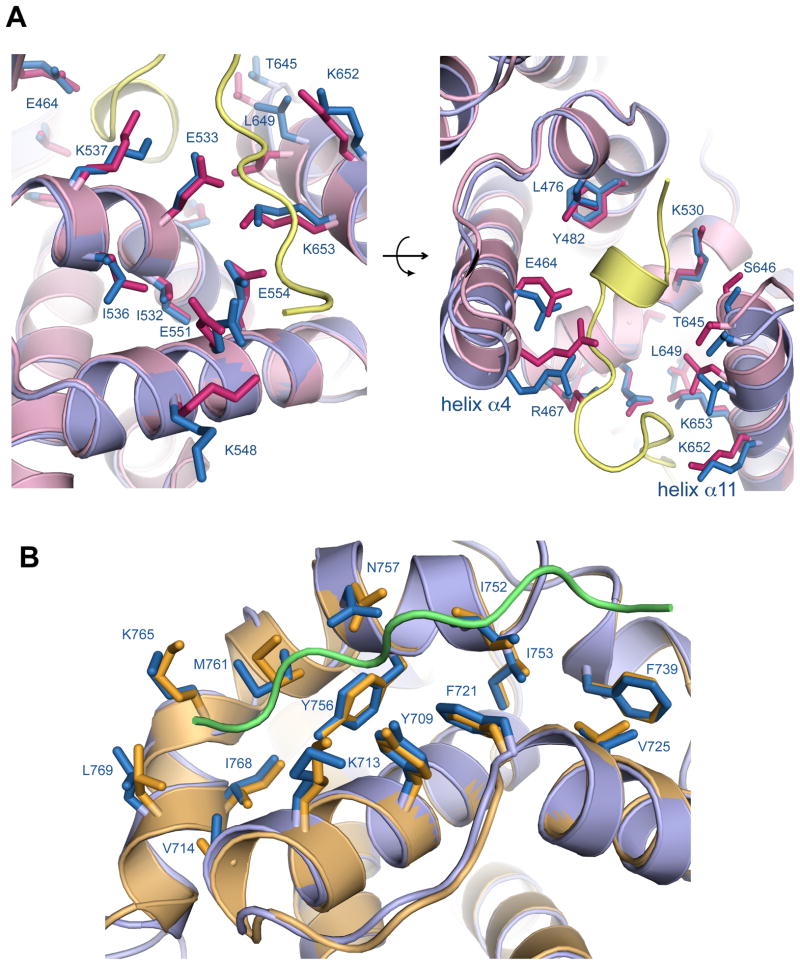
Examination of the E2FTD binding site reveals several differences in sidechain positions between the unliganded and E2FTD bound structures [Fig. 2(A)]. These changes occur such that the sidechains can make specific contacts with the E2FTD peptide. For example, K548 in Rb adopts a different conformation to make salt bridges with D410 and D411 in E2FTD. E464 in Rb moves to form a hydrogen bond with the Ser 423 sidechain in E2FTD, and R467 moves to form a salt bridge with Asp424 in E2FTD. The last two turns of helix α4 in the E2FTD-bound Rb structure are consequently tilted relative to the unliganded structure. The first two turns of helix α11 are similarly tilted towards the peptide as both T645 and L649 move to make van der Waals contacts. The rmsd for the E2FTD binding residues in the two pocket structures is 0.81 Å (comparing all atoms). This magnitude of structural change is typical for induced-fit ligand binding interactions according to a recent survey of 60 enzymes in the Protein Data Bank.15
Figure 2.
Structural changes in the pocket domain binding sites upon ligand association. (A) Close-up view of the E2FTD binding site. The aligned unliganded Rb (blue) and E2FTD-bound (pink) structures are shown along with the E2FTD peptide (yellow). (B) Close-up view of the LxCxE binding site. The aligned unliganded Rb (blue) and E7-bound (gold) structures are shown along with the E7 LxCxE peptide (green).
Fewer differences are observable at the LxCxE binding site in comparing the unliganded structure to the structure with the E7-bound peptide [Fig. 2(B)]. Association at this interface is primarily stabilized through insertion of three sidechains from the LxCxE peptide into pockets formed by hydrophobic residues from Rb. The positions of the Rb sidechains that create the binding pockets are nearly identical in both the unbound and bound structures (rmsd of 0.47 Å for all atoms in the binding residues). The crystallographic temperature factors of these sidechains are already relatively low in the unliganded structure and remain low upon binding the E7 peptide [Fig. 1(C)]. These observations suggest that compared to the E2FTD binding site, the LxCxE binding site is more preformed in the unliganded structure.
Overall the structural comparison emphasizes that the pocket domain is a rigid molecule unaltered by protein-protein interactions. The lack of allosteric changes supports observations that binding of E2F and proteins that access the LxCxE site are independent.5 It has been suggested that upon phosphorylation, RbC binds the pocket at the LxCxE site and exposes S567 through a considerable restructuring of the domain; subsequent S567 phosphorylation is proposed to cause E2F dissociation by destabilizing the hydrophobic core of the pocket.12 Although a crystal structure of the phosphorylated RbC-pocket complex has not been solved, such a mechanism seems improbable considering the lack of ligand induced structural changes observed here. Alternatively, we suggest that models for E2F dissociation in which phosphorylated Rb sequences compete directly with E2F and ‘LxCxE’ proteins at rigid pocket binding sites are more plausible in light of the available structural data.11,13
Methods
The human Rb protein from residues 380–787 was expressed in E. coli as a GST-fusion protein. The fusion protein was first purified using glutathione affinity chromatography. The GST tag was then cleaved with TEV protease, and the pocket domain was purified further with heparin chromatography. The protein was concentrated to ~30 mg/mL and injected onto a Superdex 200 column equilibrated in a buffer containing 25 mM Tris, 200 mM NaCl, and 5 mM DTT (pH=8.0). The eluted protein at a concentration of ~20 mg/mL was used directly in crystallization trials without further concentration. Crystals were grown in sitting drops at 4°C by mixing protein solution in a 1:1 ratio with well solution containing 100 mM CAPS and 10% PEG 3350 (pH=10.6). Crystals were harvested by transferring to a solution containing 100 mM CAPS, 19% PEG 3350, 10% ethylene glycol, and 12.5% sucrose (pH=10.6) and then flash freezing in liquid nitrogen.
Diffraction data were collected on Beamline 7.1 at the Stanford Synchrotron Radiation Lightsource. Diffraction spots were integrated with Mosflm16 and scaled with SCALE-IT.17 Phases were solved by molecular replacement using PHASER18 and the structure of Rb bound to the CR1 domain of E7 (PDB ID: 2R7G) as a search model. The initial model was rebuilt with Coot19 and was refined with Phenix.20 Several rounds of position refinement with simulated annealing and individual temperature factor refinement with default restraints were applied. Pairwise structure alignments were calculated with PyMol (www.pymol.org). Sidechain positions for Rb residues composing the ligand binding sites were confirmed in the final model by generating omit (Fo Fc) electron density maps.
Supplementary Material
Acknowledgments
The authors acknowledge the staff of beamline 7.1 at the Stanford Synchrotron Radiation Lightsource for assistance with data collection. This work is supported by grants from the National Institutes of Health (R01CA132685) to S.M.R and from the Department of Energy (DOE) Integrated Diffraction Analysis (IDAT) grant contract number DE-AC02-05CH11231 for SAXS data collection at the Advanced Light Source. E.M.B. was supported by an NIH training grant (T32GM008646). S.M.R. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts.
References
- 1.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8(9):671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 3.Macaluso M, Montanari M, Giordano A. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene. 2006;25(38):5263–5267. doi: 10.1038/sj.onc.1209680. [DOI] [PubMed] [Google Scholar]
- 4.Zheng L, Lee WH. The retinoblastoma gene: a prototypic and multifunctional tumor suppressor. Exp Cell Res. 2001;264(1):2–18. doi: 10.1006/excr.2000.5129. [DOI] [PubMed] [Google Scholar]
- 5.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 6.Kim HY, Ahn BY, Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. Embo J. 2001;20(1–2):295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C, Chang JH, Lee HS, Cho Y. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 2002;16(24):3199–3212. doi: 10.1101/gad.1046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391(6670):859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007;21(21):2711–2716. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao B, Spencer J, Clements A, Ali-Khan N, Mittnacht S, Broceno C, Burghammer M, Perrakis A, Marmorstein R, Gamblin SJ. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc Natl Acad Sci U S A. 2003;100(5):2363–2368. doi: 10.1073/pnas.0436813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem. 285(21):16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 13.Rubin SM, Gall AL, Zheng N, Pavletich NP. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123(6):1093–1106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 14.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutteridge A, Thornton J. Conformational changes observed in enzyme crystal structures upon substrate binding. J Mol Biol. 2005;346(1):21–28. doi: 10.1016/j.jmb.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 17.Howell PL, Smith GD. Identification of Heavy-Atom Derivatives by Normal Probability Methods. Journal of Applied Crystallography. 1992;25:81–86. [Google Scholar]
- 18.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.