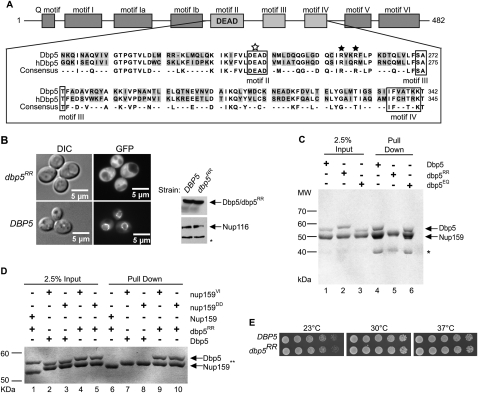
Figure 3.
dbp5RR mutant does not bind Nup159 and displays no in vivo growth defects. (A) Amino acid residue alignment for a portion of S. cerevisiae Dbp5 and Homo sapiens hDBP5. Black stars designate conserved arginine residues that were changed to aspartic acid residues (R256D and R259D) in Dbp5 to yield dbp5RR. The open white star represents a conserved glutamate residue that was changed to glutamine (E240Q) to yield dbp5EQ. Conserved DEAD-box family sequence motifs are boxed and identified below alignment. (B) Cytoplasmic mislocalization of GFP-dbp5RR by live-cell microscopy: S. cerevisiae strains with DBP5 chromosomally deleted, expressing either GFP-DBP5 or GFP-dbp5RR on plasmids, were used in live-cell fluorescent microscopy experiments to visualize subcellular localization of each protein. (Left) DIC image. (Right) GFP. Western blot analysis of yeast whole-cell extracts was conducted to test for relatively equal levels of expression of GFP-Dbp5 and GFP-dbp5RR. Nup116 protein level was used as a loading control. (*) Degradation products of Nup116. (C) Nup159-NTD interaction in vitro with dbp5RR is not detected: Soluble binding assays were conducted with 200 pmol of bacterially expressed recombinant His6-Nup159-NTD and immobilized on Ni-NTA agarose resin and either recombinant Dbp5, dbp5RR, or dbp5EQ. Input and bound fractions were analyzed by SDS-PAGE and Coomassie staining. (*) Nup159-NTD degradation product. (D) Swapping the salt bridge interaction allows nup159DD and dbp5RR interaction: Soluble binding assays were conducted as in C with His6-Nup159-NTD, His6-nup159VI-NTD, His6-nup159DD-NTD, Dbp5, or dbp5RR. (E) dbp5RR strain does not have growth defects: S. cerevisiae strains with DBP5 chromosomally deleted, expressing either wild-type DBP5 or dbp5RR, were used to perform serial dilution growth assays at 23°C, 30°C, and 37°C.