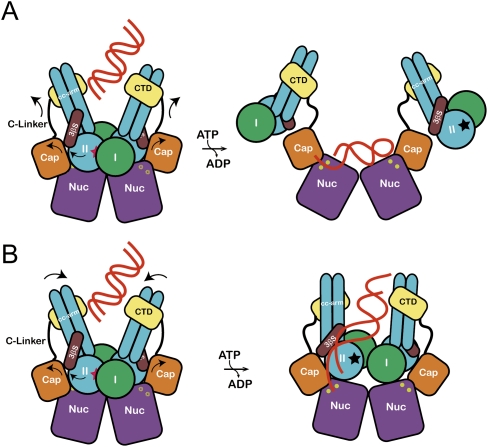
Figure 7.
The proposed allosteric regulation models for the MjMR complex. (A) DNA binds to the central groove and stimulates the ATP hydrolysis. Rotation of lobe II (an arrow) triggers the movement of the capping domain and C-terminal domain of MjMre11, which dislocates MjRad50CD and unmasks the active site (yellow circle) of MjMre11. The DNA-binding mode is similar to that of the PfMre11–DNA complex. A dark star represents an ATP-binding site in MjRad50. (B) Rotation of lobe II of MjRad50 is accompanied by movement of the C-terminal domain of MjMre11 and induces the rotation of coiled-coils of MjRad50 such that their positions changed from V shape to parallel (an arrow). This conformational change may open the active site channel to comfortably accommodate the DNA substrate. The DNA molecule bound to the groove passes through a three-stranded sheet (β9–β11) and is guided to the active site channel.