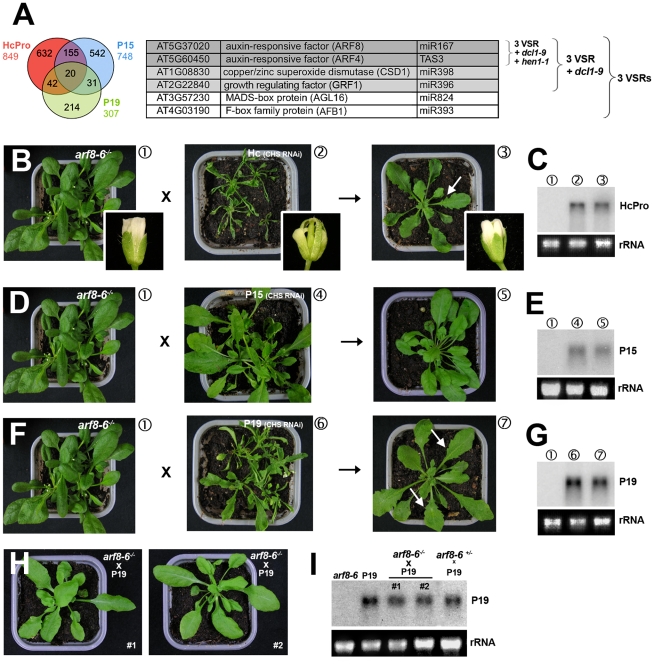
Figure 4. Heterozygous and homozygous arf8 mutant backgrounds respectively attenuate and alleviate the developmental phenotypes incurred by VSRs.
(A) The Venn diagram on the left shows that only a modest number of transcripts are up-regulated in common in leaves of the three VSR transgenics. The table shows that refining the analysis with additional filters based on transcripts up-regulated in dcl1-9 (pale grey) and hen1-1 (dark grey) backgrounds singularizes ARF4 and ARF8, respectively direct targets of miR390 and miR167, as strong candidates for the underlying developmental defects seen in VSR transgenics. (B–C) Strong reduction of leaf and inflorescence defects (inlays) caused by HcPro in F1 progenies of crosses between arf8 mutants and HcPro transgenics carrying the CHS RNAi transgene (B). The Northern blot in (C) shows comparable accumulation of HcPro transcripts in the various backgrounds involved in the crosses. (D–E) same as (B–C) for P15 transgenics with the CHS RNAi background. (F–G) Same as (B–C) for P19 transgenics with the CHS RNAi background. Arrows indicate the presence of slight leaf serration in F1 progeny plants. (H–I) Complete alleviation of developmental defects and sterility of P19 transgenic plants (CHS RNAi background) in the homozygous arf8 mutant background. Northern analysis in (I) confirms comparable P19 levels in the various backgrounds indicated. Plants #1 and #2 where retrieved through independent genotyping in populations of P19 plants with homozygous or heterozygous arf8 mutant genotype. rRNA: ethidium bromide staining of ribosomal RNA provides a control for equal RNA loading.